Can I use the FlowClean R Script with FCS Express?
Yes, with FCS Express 6 or later, you can run the FlowClean R Script via the R Integration.
About FlowClean
FlowClean is an algorithm and tool for quality control, designed to identify fluorescence abnormalities caused by fluidic issues at the time of acquisition. FlowClean tracks subset frequency changes within a sample during acquisition, and "flags" time periods with fluorescence perturbations that could possibly lead to the emergence of false populations. The result of the FlowClean R script, when used in FCS Express, is a new parameter, called FCSE_GoodVsBad. The end user can utilize the newly-created parameter to exclude events within aberrant time periods.
The FlowClean algorithm can be run from within the R Integration tool of FCS Express 6 or later by using an adapted version of the original (flowClean_1.12.0) code. For simplicity, the original script has been adapted to use a matrix as input parameter instead of a standard FCS file, while the remaining portions of the code and algorithm are unchanged.
Note: the original FlowClean script works on raw fluorescence data, which are uncompensated and not transformed (i.e. linear) for FCS 3.0 and 3.1 files. However, the matrix exported by FCS Express into R reflects what is displayed in the plot to which the transformation has been applied. Since compensated data are usually displayed in the plots, these compensated data will be exported to R instead of uncompensated data. The results obtained when FlowClean is run through FCS Express will not reflect the results obtained when FlowClean is run directly through R. The same consideration needs to be taken into account with regards to parameter scaling. By default, FCS Express exports fluorescence parameters with biexponential scaling. When this is the case, the results obtained when FlowClean is run through FCS Express may not reflect the results obtained when FlowClean is run directly through R. The scale applied to parameters when exported to R can be customized (e.g. set to linear) in the Transformations dialog.
For more details about the modifications made to the original script in order to be run through FCS Express, please see the comments within the script or contact us at support@denovosoftware.com.
Please Note: The FlowClean R implementation may fail on a wide variety of data files due to the algorithm itself failing. Please use this tool at your own risk.
How to Use the FlowClean R Script
FlowClean makes use of additional R packages that must be installed via R prior to using in FCS Express.
- To install the FlowClean package, please run the following lines in your R console:
source("https://bioconductor.org/biocLite.R")
biocLite("flowClean")
- To confirm the installation was performed properly, please load the FlowClean package in your R console by running the following line:
library("flowClean")
- The following lines should be returned, which verifies the package was installed properly:
> library("flowClean")
Loading required package: flowCore
>
If any errors or problems are encountered, please contact us at support@denovosoftware.com.
FlowClean makes use of additional R packages that must be installed via R prior to using in FCS Express.
- To install the FlowClean package, please run the following lines in your R console:
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("flowClean")
- To confirm the installation was performed properly, please load the FlowClean package in your R console by running the following line:
library("flowClean")
- The following lines should be returned, which verifies the package was installed properly:
> library("flowClean")
Loading required package: flowCore
>
If any errors or problems are encountered, please contact us at support@denovosoftware.com.
FlowClean can be run in FCS Express via the steps below. Please ensure the R Application Bridge is set to be open in your FCS Express Options.
- Open FCS Express.
- Load a data file into the layout and create a plot.
- Create a new Transformation for R Add Parameters (please refer to the FCS Express manual to learn how to create an R Add Parameter transformation).
- Choose the FlowClean_FCSE.R script as the R Script File in the Transformations dialog.
- Check all of the Fluorescence parameters and the time parameter in the parameter list check box section. Note that Scatter parameters are generally not included.
- Select Events as row in the Events order section of the Transformations dialog.
- Drag the new Transformation from step 3 from the Transformation dialog onto the plot you inserted in step 2.
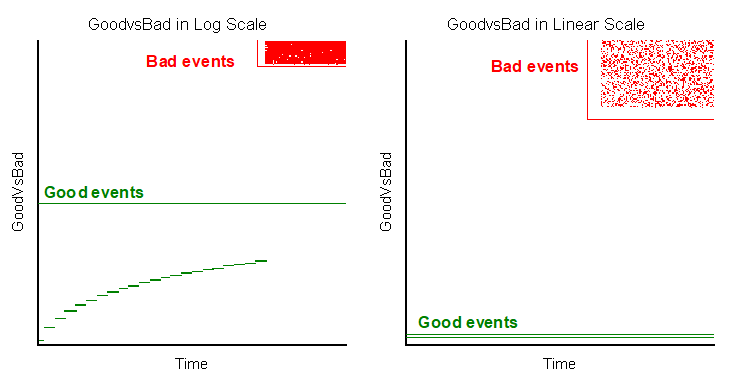
The result of the FlowClean algorithm will be now available as an additional parameter called FCSE_GoodVsBad that will allow the discrimination between “Good” and “Bad” events, where “Bad” means perturbations caused by the fluidics. The algorithm will assign a numeric value to each event. When default settings of the script are used, numbers from 1 to 100 are assigned to Good events while numbers from 10000 to 20000 are assigned to Bad events. Once the FCSE_GoodVsBad parameter is available, gates can be drawn around Good events (to include them in the downstream analysis) or around Bad events (to exclude them from the downstream analysis) as depicted below:
However, there are exceptions to this behavior outlined in the following section.
- When no time parameter is available, the GoodVsBad parameter will contain only 0 (zero).
- When the number of events is lower than 30000, the GoodVsBad parameter will contain only 0 (zero).
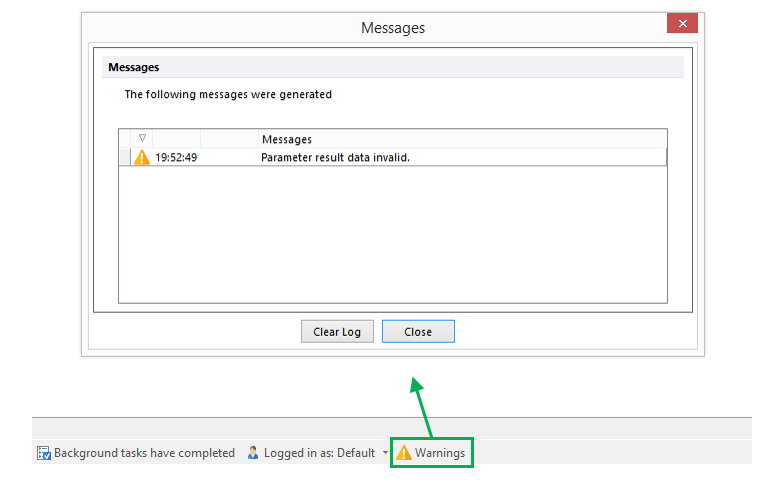
- When the script encounters a fault or error due to the FlowClean algorithm failing, no parameter will be created and a warning message will be reported in FCS Express. See example below:
-
Installation
-
Licenses
-
- Can I get more information regarding the Add-Ons that can be purchased with a license?
- Can I lock my template based on an electronic signature?
- Does FCS Express have any features to help meet 21 CFR Part 11 compliance?
- Does FCS Express have Quality Control features?
- Does FCS Express offer Single Sign On capability?
- How do I configure SQL Server to host a database for FCS Express?
- What database options are available when I purchase the Security option?
- What is the difference between the different types of Users that are available with a Security and Logging license?
- What is the difference between the Logging option and System Level Audit Trails?
- What SQL Server permissions are needed?
-
-
- Can I share my USB dongle or countercode license with another user?
- Can I track usage of the internet dongle?
- Can I try out the Internet Dongle before I make a purchase?
- Can the administrator log users out?
- Do you have to be connected to the internet at all times with the Internet dongle?
- How can users be added to an internet dongle license?
- How do I activate my dongle?
- How do I change my internet dongle/site license password?
- How many people can be logged in at the same time?
- How many user accounts can I create?
- If a user left the computer running can the user log themselves out from another computer?
- What are the differences between the internet dongle and network licensing options?
- What happens if I lose my internet connection?
- What happens if the user leaves the computer without logging out?
- What happens to the users login in case of an unexpected interruption? For instance, a software crash, power failure, etc.
- Why am I receiving a message that FCS Express cannot connect to De Novo Software servers?
- Show all articles ( 1 ) Collapse Articles
-
-
- Can I convert my Cytek license from the countercode licensing option to another licensing option?
- How can I claim my license purchased through BD Accuri Cytometers?
- How can I claim my license purchased through BD Biosciences?
- How can I claim my license purchased through Nexcelom Biosciences?
- How can I claim my license purchased through Sysmex-Partec GmbH?
- How can I claim the FCS Express license that came with my Cytek instrument purchase?
-
-
Layouts & Loading Data
-
- Can I customize my Quick Access Toolbar?
- For an existing layout, can I change the way FCS Express remembers parameters?
- How can I exclude histogram overlays from batch processing?
- How do I change my layout from portrait to landscape?
- How do I change the resolution of my exported images?
- How do I re-establish data files that were linked to a layout?
-
- Are Beckman Coulter LMD files unique?
- Can I find a support resource page for the analysis of Cytek data in FCS Express?
- How can I easily create the "filename" column in the "ExtraKeywords Table.csv" file?
- How can I load a Sony MA900 Index Sort file into FCS Express?
- How can I load data from the BD Accuri C6 Flow Cytometer?
- How can I load MACSQuantify files that were exported from MACSQuantify software version 3.0.1?
- How can I quickly reload all of the data files in the data list?
- How do I change the display in my plots from one data file to another data file?
- How do I export .ICE files from Thermo Cellomics HCS Studio?
- How do I tell FCS Express what plate size to use if that information is not included in the data file?
- How do I upload files to the De Novo Software FTP site?
- How do I use BD Accuri CFlow files with Multicycle DNA analysis in FCS Express?
- How do I work with images from the Thermo Scientific Attune CytPix?
- What is the Elapsed Time setting in the Gallios software and how do I convert it to real time?
- Why am I seeing a warning message when loading my Cytek data onto a layout object?
- Why are iterations in my Data List gray?
- Why are there sometimes access violations when I save and load files?
- Why do I get the message that a data file exported from a FACSDiva™ Experiment is invalid?
- Show all articles ( 3 ) Collapse Articles
-
- How are existing quadrants handled when an old layout is opened in version 7.20 and later?
- How can I set quadrants to behave like conventional gates?
- How can I set quadrants to behave like in earlier versions?
- How can quadrants be linked?
- Quadrants in FCS Express versions 7.20 / 7.24 and later
- Why does the Quadrants Options window appear when I open an older layout in version 7.24?
- Why have percentages reported by quadrants changed after updating to FCS Express version 7.20.20?
-
-
Data Analysis
-
- Caveats when using Biexponential Scaling with automatic Below Zero parameter detection in the presence of outliers.
- How can I create a merged data with equally-sized downsampled samples?
- How can I do pre-processing for high-dimensional data analysis?
- How can I explore tSNE/UMAP plots?
- How do I use SPADE?
- What is FlowSOM?
- What is T-SNE?
- What is UMAP?
-
- Can FCS Express integrate Python scripts?
- Can I use the FlowAI script in FCS Express?
- Can I use the FlowClean R Script with FCS Express?
- How can I create a matrix of autofluorescence, and import autofluorescence into EasyPanel?
- How can I recreate ratiometric data acquired in FACSDiva?
- How do I use R Integration with FCS Express?
- How does FCS Express implement software compensation?
- If my data does not have a Time parameter, can I create one?
- What is compensation?
- What is the compensation workflow in FCS Express?
- When acquiring spectral data, should my single-stained controls be "as bright or brighter" than my fully-stained sample?
-
- Can a set of quadrants be both percentile and floating?
- Can I customize the display of my data from different instruments?
- Can I disable the live updating feature?
- How can I display all of my detectors for my Cytek data?
- How can I set FCS Express so my FCS 3.0 biexponential data looks the same as it did in the BD FACSDiva software?
- How do I display Summit data in FCS Express as it appears in the Summit Software?
- How do I fix the biexponential axes on a plot?
- How do I rescale CytoFLEX data so it displays as it did at acquisition?
- How do I update my density and contour plots created in Version 4 to use the newest color palette?
- What are resolution options?
- What is Biexponential and Hyperlog Scaling?
- What is the best way to set FCS Express to display FCS 3.0 data from FACSDiva on a 4 decade log scale?
- Where can I get more information regarding DNA analysis using the Multicycle AV?
- Why can’t I change my plot axis labels from the Name keyword to the Stain keyword?
- Why do I see a warning message when inserting a Spectrum Plot?
- Why do my dot plots appear sparse and blocky?
- Why is the text on the right most label cut off my plot?
- Show all articles ( 2 ) Collapse Articles
-
- Can I create an output file that contains the same plot from each data file on a single page?
- Can I export spectral data to the FCS format?
- How can I successfully export a GatingML file?
- How do the batch processing run modes differ, and why would I use them?
- Why do I get an “Old format or invalid type library” error when using Microsoft excel during batch analysis?
-
- How are statistics in FCS Express calculated compared to how they are calculated in BD FACSDiva?
- How can I display my statistical data in Scientific Notation?
- How do I calculate EC/IC Anything?
- What is “Stain Index” and how do I calculate it with FCS Express?
- What is MFI (Mean or Median Fluorescence Intensity) and how do I calculate it in FCS Express?
- Why have percentages reported by quadrants changed after updating to FCS Express version 7.20.20?
- Why is the Geometric Mean being reported as NaN or ##ERROR##?
-
-
Image Cytometry
-
- How do I adjust the axes to display small particle data from Amnis CellStream?
- How do I choose which images and parameters to view in a Data Grid?
- How do I export/save data from IDEAS software and load it in FCS Express?
- How do I make my images in the data grid larger?
- How do I pseudo-color images in a data grid?
- How do I work with Amnis derived image cytometry data in FCS Express?
-
- Can I display heat maps with my Image Cytometry data?
- Can I work with data from PerkinElmer Instruments?
- Do you offer 21 CFR Part 11 compliance options for the Image Cytometry Version?
- Do you offer image segmentation or image analysis?
- How can Attune™ CytPix data sets with images (.ACS files) be merged for high dimensional analysis?
- How do I use CellProfiler Data with FCS Express?
- How do I use ImageJ with FCS Express?
- What file formats are compatible with FCS Express Image Cytometry?
- Where can I find Nexcelom Resources and Applications?
-
-
FCS Express on Mac
-
Upgrading FCS Express
-
- Can different versions of FCS Express exist on the same computer?
- How can I view and convert my V3 layouts to FCS Express 7?
- How do I import my version 3 security databases into newer versions of FCS Express?
- How do I update Density Plots created in Version 4?
- Is there an upgrade discount from earlier versions of FCS Express?
- Version 4 Internet Dongle Retirement
- Why are my density plots from V3 not displayed correctly in later versions?
- Why are there fewer outlier dots on my FCS Express 5 and later density plots than in V4?
-
Clinical & Validation Ready
-
- Can I get more information regarding the Add-Ons that can be purchased with a license?
- Can I lock my template based on an electronic signature?
- Does FCS Express have any features to help meet 21 CFR Part 11 compliance?
- What is the difference between the different types of Users that are available with a Security and Logging license?
- What is the difference between the Logging option and System Level Audit Trails?
 Explore the Scientific R&D Software
Explore the Scientific R&D Software