How do I use SPADE?
High-dimensional single-cell technologies, such as Flow, Mass and Image cytometry, can measure dozens of parameters at the single-cell level. FCS Express integrates Spanning-tree Progression Analysis of Density-normalized Events, otherwise known as SPADE, which is a tool that extracts a hierarchy from high-dimensional cytometry data in an unsupervised manner and allows users to visualize multiple cell types in a branched tree structure without requiring the user to define a known cellular ordering.
The final result of the algorithm in FCS Express is a Heat Map plot in which each cell type is depicted as a node of the branched tree. The Heat Map plot can be formatted to color each node based on the expression of a given marker. Moreover, the size of each node can be made proportional to a given statistic (e.g., the number of events within the node).
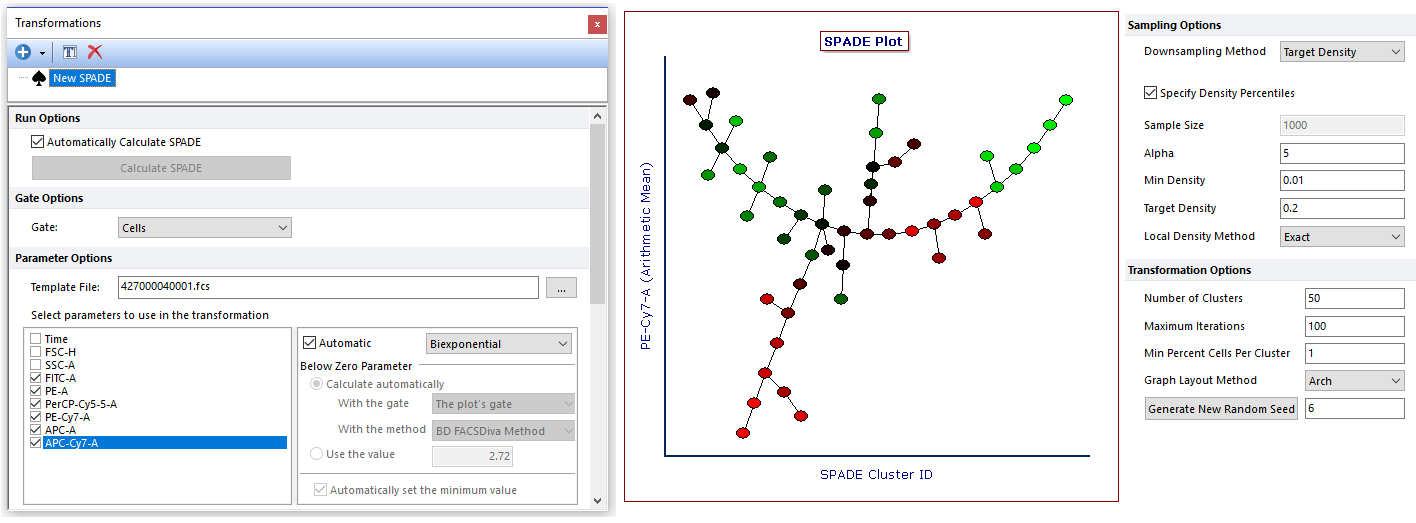
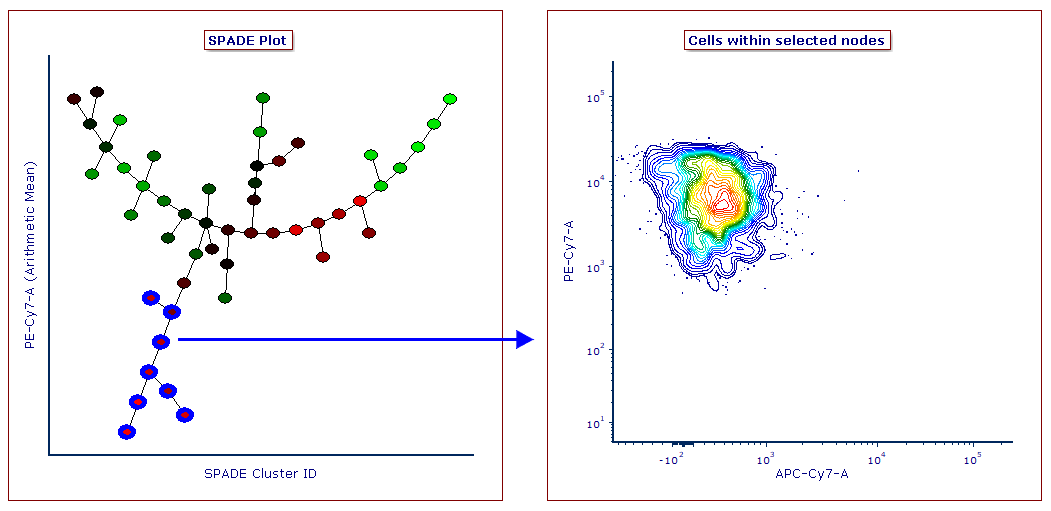
Gates may be defined on SPADE transformed Heat Map plot to select one or multiple nodes for downstream analysis. Similarly, gates created on classical dot plots can be applied to SPADE plots as well.
Learn more about using SPADE via the FCS Express Manual at:
SPADE (Spanning-tree Progression Analysis of Density-normalized Events) is a clustering algorithm. Its goal is to automatically identify cell populations in multidimensional cytometry data files. FCS Express 6 and later allows users to run SPADE on Flow, Mass and Image cytometry data.
More details on SPADE can be found in the following publications:
SPADE and k-Means are examples of clustering algorithms. The goal of the algorithms is to group data (i.e., cells) together in a way that data in the same group (i.e., the same cluster) are more similar to each other than data from other groups (i.e., other clusters). With respect to k-Means, SPADE includes a series of computational steps that:
- Allow rare events to form their own clusters instead of being outnumbered by abundant cell populations during clustering.
- To generate a minimum spanning tree representation in which clusters are connected with minimum total edge length. t-SNE and PCA are examples of dimensionality reduction algorithms. A dimensionality reduction algorithm attempts to map high-dimensional data into lower dimensions by finding a data set in low-dimensional space that, in some way, represents the original high-dimensional data.
There are many reasons to choose FCS Express to run the SPADE transformation:
- The user interface that allows users to run SPADE in FCS Express is very intuitive and easy to use.
- SPADE analysis in FCS Express is completely integrated with the layout, meaning that the user can draw gates on SPADE plots to select one or multiple nodes for downstream analysis. Similarly, gates drawn on classical dot plots can be applied to SPADE plots as well.
- SPADE is extremely fast in FCS Express, as the team at De Novo Software made great efforts to enhance running of the algorithm and the software in general.
Check out our recorded webinar below to see why you should be Using SPADE and data transformation tools in FCS Express!
-
Installation
-
Licenses
-
- Can I get more information regarding the Add-Ons that can be purchased with a license?
- Can I lock my template based on an electronic signature?
- Does FCS Express have any features to help meet 21 CFR Part 11 compliance?
- Does FCS Express have Quality Control features?
- Does FCS Express offer Single Sign On capability?
- How do I configure SQL Server to host a database for FCS Express?
- What database options are available when I purchase the Security option?
- What is the difference between the different types of Users that are available with a Security and Logging license?
- What is the difference between the Logging option and System Level Audit Trails?
- What SQL Server permissions are needed?
-
-
- Can I share my USB dongle or countercode license with another user?
- Can I track usage of the internet dongle?
- Can I try out the Internet Dongle before I make a purchase?
- Can the administrator log users out?
- Do you have to be connected to the internet at all times with the Internet dongle?
- How can users be added to an internet dongle license?
- How do I activate my dongle?
- How do I change my internet dongle/site license password?
- How many people can be logged in at the same time?
- How many user accounts can I create?
- If a user left the computer running can the user log themselves out from another computer?
- What are the differences between the internet dongle and network licensing options?
- What happens if I lose my internet connection?
- What happens if the user leaves the computer without logging out?
- What happens to the users login in case of an unexpected interruption? For instance, a software crash, power failure, etc.
- Why am I receiving a message that FCS Express cannot connect to De Novo Software servers?
- Show all articles ( 1 ) Collapse Articles
-
-
- Can I convert my Cytek license from the countercode licensing option to another licensing option?
- How can I claim my license purchased through BD Accuri Cytometers?
- How can I claim my license purchased through BD Biosciences?
- How can I claim my license purchased through Nexcelom Biosciences?
- How can I claim my license purchased through Sysmex-Partec GmbH?
- How can I claim the FCS Express license that came with my Cytek instrument purchase?
-
-
Data Analysis
-
- Can FCS Express integrate Python scripts?
- Can I use the FlowAI script in FCS Express?
- Can I use the FlowClean R Script with FCS Express?
- How can I recreate ratiometric data acquired in FACSDiva?
- How do I use R Integration with FCS Express?
- How does FCS Express implement software compensation?
- If my data does not have a Time parameter, can I create one?
- What is compensation?
- What is the compensation workflow in FCS Express?
- When acquiring spectral data, should my single-stained controls be "as bright or brighter" than my fully-stained sample?
-
- Can a set of quadrants be both percentile and floating?
- Can I customize the display of my data from different instruments?
- Can I disable the live updating feature?
- How can I display all of my detectors for my Cytek data?
- How can I set FCS Express so my FCS 3.0 biexponential data looks the same as it did in the BD FACSDiva software?
- How do I display Summit data in FCS Express as it appears in the Summit Software?
- How do I fix the biexponential axes on a plot?
- How do I rescale CytoFLEX data so it displays as it did at acquisition?
- How do I update my density and contour plots created in Version 4 to use the newest color palette?
- What are resolution options?
- What is Biexponential and Hyperlog Scaling?
- What is the best way to set FCS Express to display FCS 3.0 data from FACSDiva on a 4 decade log scale?
- Where can I get more information regarding DNA analysis using the Multicycle AV?
- Why can’t I change my plot axis labels from the Name keyword to the Stain keyword?
- Why do my dot plots appear sparse and blocky?
- Why is the text on the right most label cut off my plot?
- Show all articles ( 1 ) Collapse Articles
-
- How are statistics in FCS Express calculated compared to how they are calculated in BD FACSDiva?
- How can I display my statistical data in Scientific Notation?
- How do I calculate EC/IC Anything?
- What is “Stain Index” and how do I calculate it with FCS Express?
- What is MFI (Mean or Median Fluorescence Intensity) and how do I calculate it in FCS Express?
- Why have percentages reported by quadrants changed after updating to FCS Express version 7.20.20?
- Why is the Geometric Mean being reported as NaN or ##ERROR##?
-
- Are Beckman Coulter LMD files unique?
- Can I find a support resource page for the analysis of Cytek data in FCS Express?
- How can I easily create the "filename" column in the "ExtraKeywordsTable.csv" file?
- How can I load data from the BD Accuri C6 Flow Cytometer?
- How can I load MACSQuantify files that were exported from MACSQuantify software version 3.0.1?
- How do I change the display in my plots from one data file to another data file?
- How do I export .ICE files from Thermo Cellomics HCS Studio?
- How do I tell FCS Express what plate size to use if that information is not included in the data file?
- How do I upload files to the De Novo Software FTP site?
- How do I use BD Accuri CFlow files with Multicycle DNA analysis in FCS Express?
- How do I work with images from the Thermo Scientific Attune CytPix?
- What is the Elapsed Time setting in the Gallios software and how do I convert it to real time?
- Why are there sometimes access violations when I save and load files?
- Why do I get the message that a data file exported from a FACSDiva™ Experiment is invalid?
-
Image Cytometry
-
- How do I adjust the axes to display small particle data from Amnis CellStream?
- How do I choose which images and parameters to view in a Data Grid?
- How do I export/save data from IDEAS software and load it in FCS Express?
- How do I make my images in the data grid larger?
- How do I pseudo-color images in a data grid?
- How do I work with Amnis derived image cytometry data in FCS Express?
-
- Can I display heat maps with my Image Cytometry data?
- Can I work with data from PerkinElmer Instruments?
- Do you offer 21 CFR Part 11 compliance options for the Image Cytometry Version?
- Do you offer image segmentation or image analysis?
- How do I use CellProfiler Data with FCS Express?
- How do I use ImageJ with FCS Express?
- What file formats are compatible with FCS Express Image Cytometry?
- Where can I find Nexcelom Resources and Applications?
-
-
FCS Express on Mac
-
Upgrading FCS Express
-
- Can different versions of FCS Express exist on the same computer?
- How are existing quadrants handled when an old layout is opened in version 7.20 and later?
- How can I view and convert my V3 layouts to FCS Express 7?
- How do I import my version 3 security databases into newer versions of FCS Express?
- How do I update Density Plots created in Version 4?
- Is there an upgrade discount from earlier versions of FCS Express?
- Version 4 Internet Dongle Retirement
- Why are my density plots from V3 not displayed correctly in later versions?
- Why are there fewer outlier dots on my FCS Express 5 and later density plots than in V4?
