Can FCS Express integrate Python scripts?
Although the popular algorithms for high-dimensional data analysis in cytometry are already embedded into FCS Express and do not require any additional external software or “plugins”, we know that analysis might require custom algorithms for some advanced cases and researchers. If your research group needs additional flexibility then FCS Express allows you to run Python scripts via the Python Transformation pipeline step.
 FCS Express can integrate Python scripts via the Python Transformation pipeline step. The Python pipeline step allows users to run their own Python scripts as part of FCS Express pipelines and work on the resulting output directly within FCS Express.
FCS Express can integrate Python scripts via the Python Transformation pipeline step. The Python pipeline step allows users to run their own Python scripts as part of FCS Express pipelines and work on the resulting output directly within FCS Express.
To set up your machine for Python transformations, please complete the following installation instructions.
Please note: in FCS Express 7.18, Python versions 3.11 and later are not yet compatible. The Python transformation cannot yet be run in the Mac version of FCS Express.
This step is required for PARC and TRIMAP, which are included in our FCSExpress Anaconda environment. It may also be required for other libraries.
- Go to this link: https://visualstudio.microsoft.com/visual-cpp-build-tools/ and click on Download Build Tools.
- Double-click on Vs_Buildtools.exe (it may be in your computer’s "Downloads" folder).
- If asked to "Allow Installer to make changes", click Yes.
- If you got a message about privacy, click Continue.
- From the Workloads tab (see screenshot below), choose Desktop development with C++.
- Under Optional components in the right pane (see screenshot below), only MSVC… and Windows 10 SDK (or Windows 11 SDK) are necessary. You may “uncheck” the others to save disk space.
- Click Install.
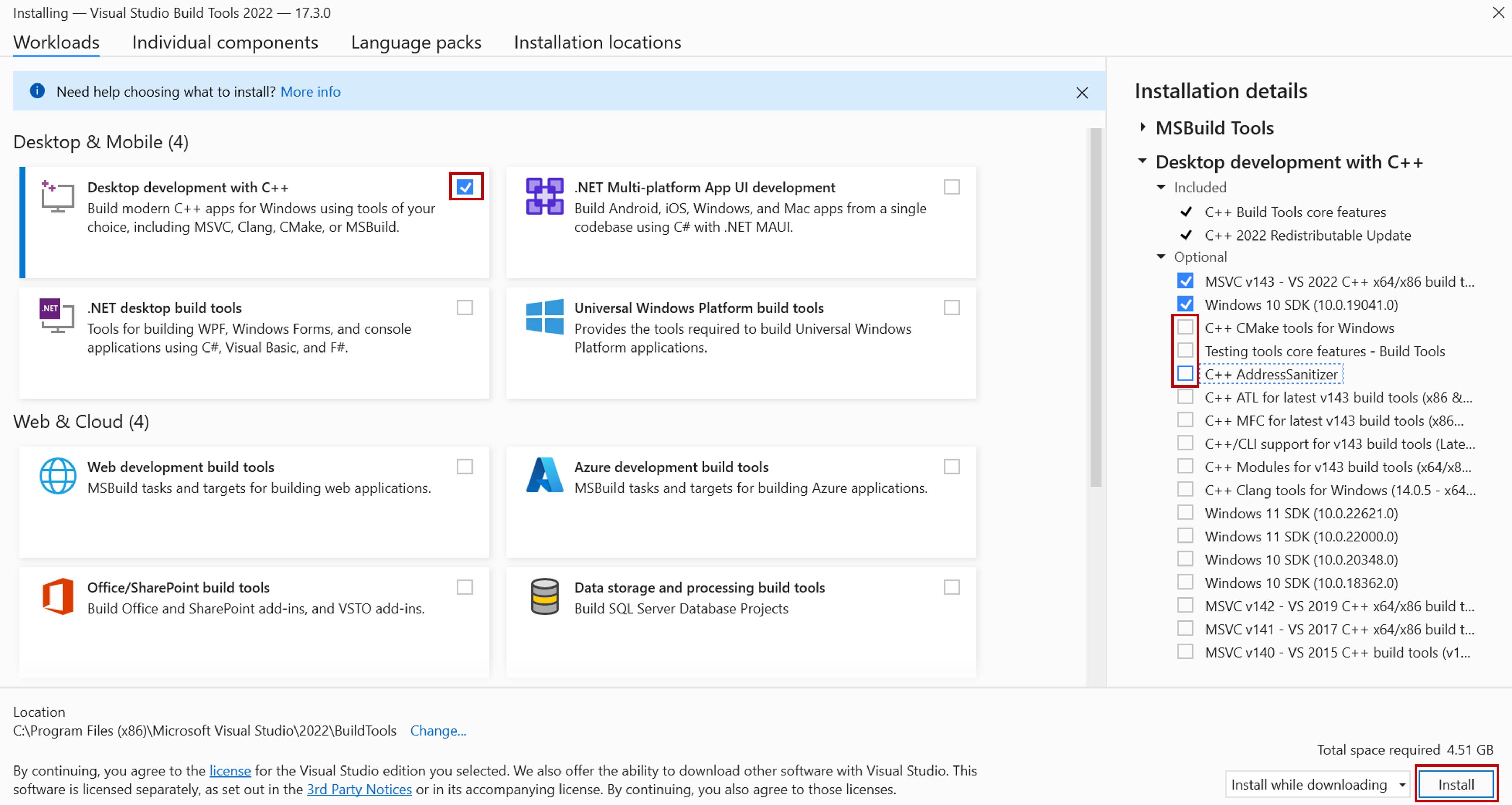
- This will take about 4 minutes to install and about 4.51 GB of space (8.15 GB on Windows 11) are required.
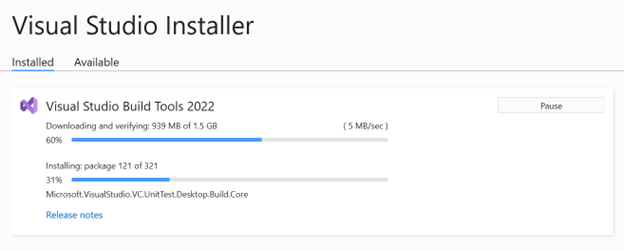
- When done, exit the Visual Studio Installer window.
- If you are prompted to restart your computer, please do so.
- You may now go on to the next step of Installing Anaconda, Python and Required Packages, found below.
Skip this step if you already have Python and the desired libraries installed on your computer.
If Anaconda is already installed on your computer, skip to the step below called "Launch Anaconda Navigator".
A convenient way to install Python is installing Anaconda, which is a distribution of Python for scientists (3.5 GB space is required).
- Follow the steps at the link below. You do not need to log in or sign up at any point when installing Anaconda and Python. If you are prompted to login or sign up, exit the pop-up window.
Once Anaconda is installed, we can create an environment to work with FCS Express. An environment is a collection of libraries of interest. Environments simplify library installations, avoid system pollution, sidestep dependency conflicts and minimize reproducibility Issues.
- Launch Anaconda Navigator from the Windows Start Menu
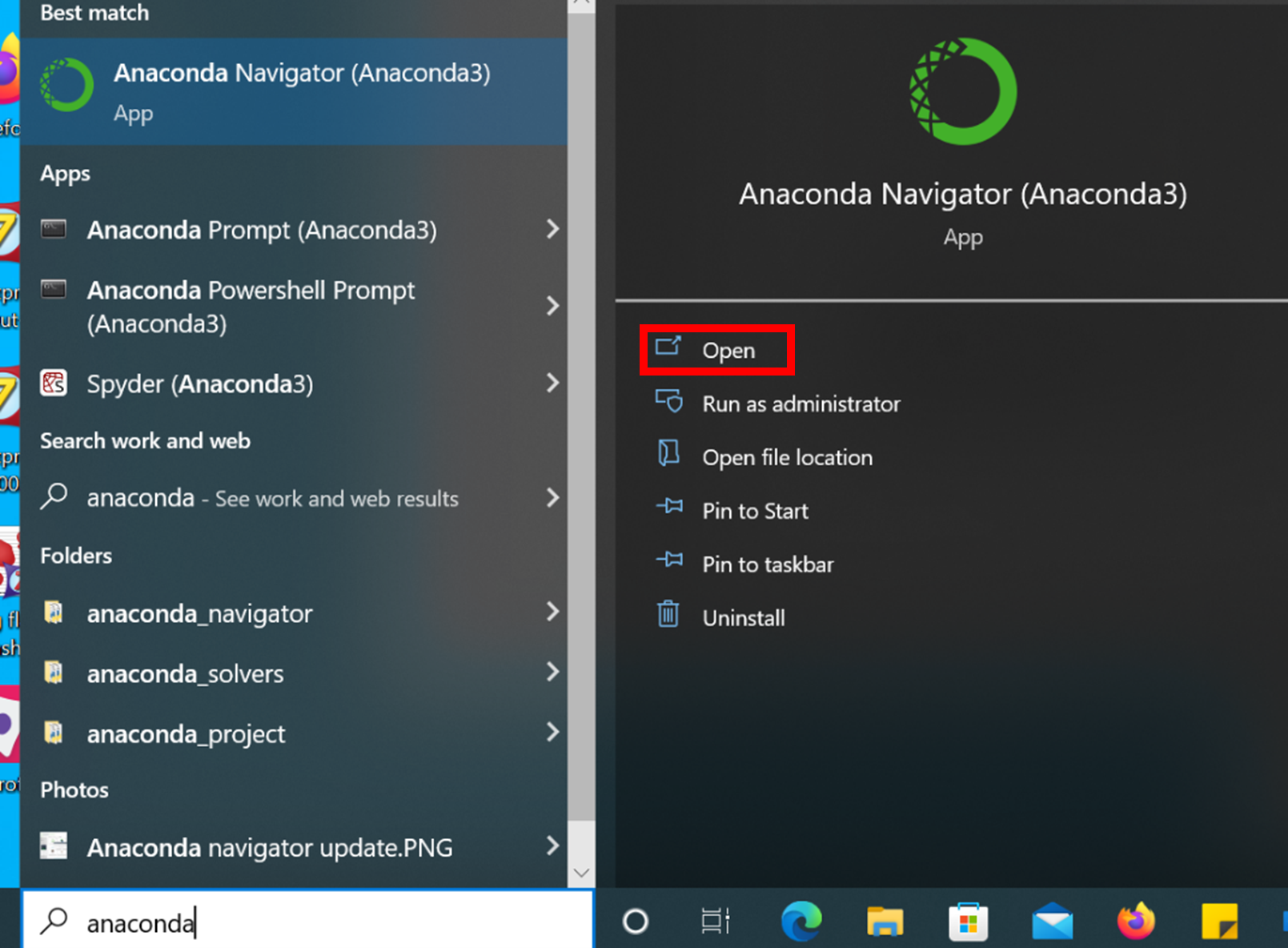
- Wait for the software to finish loading (wait for the blue animation to cease).

- If Anaconda asks to update, click Yes.
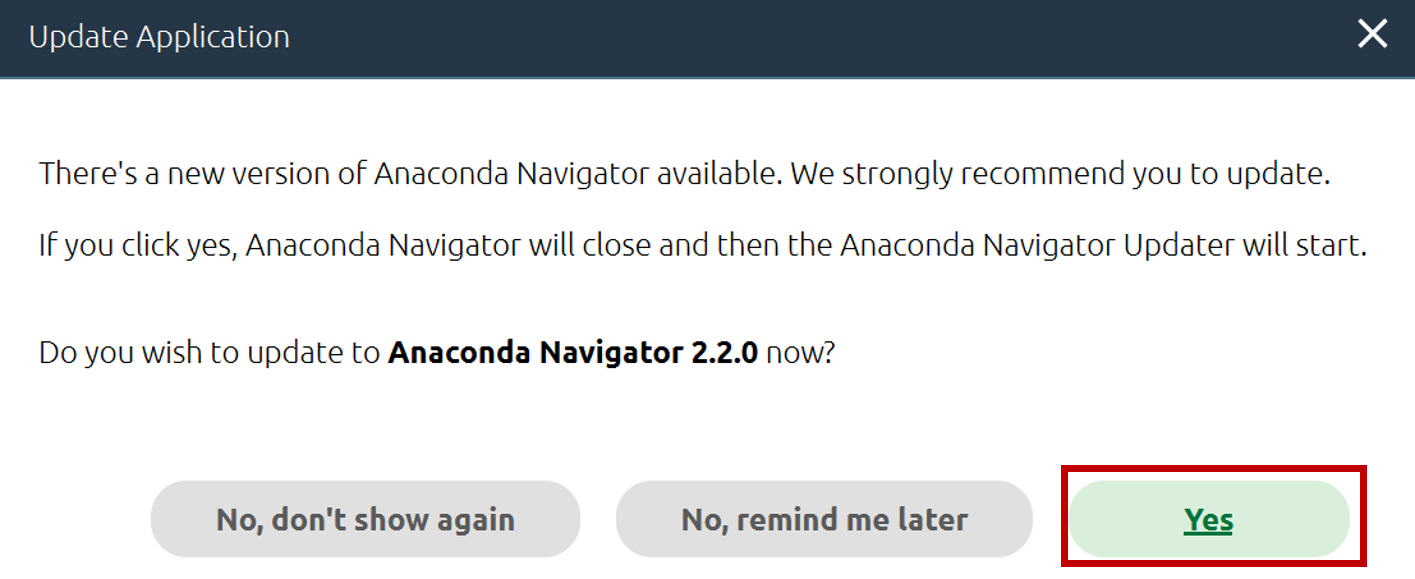
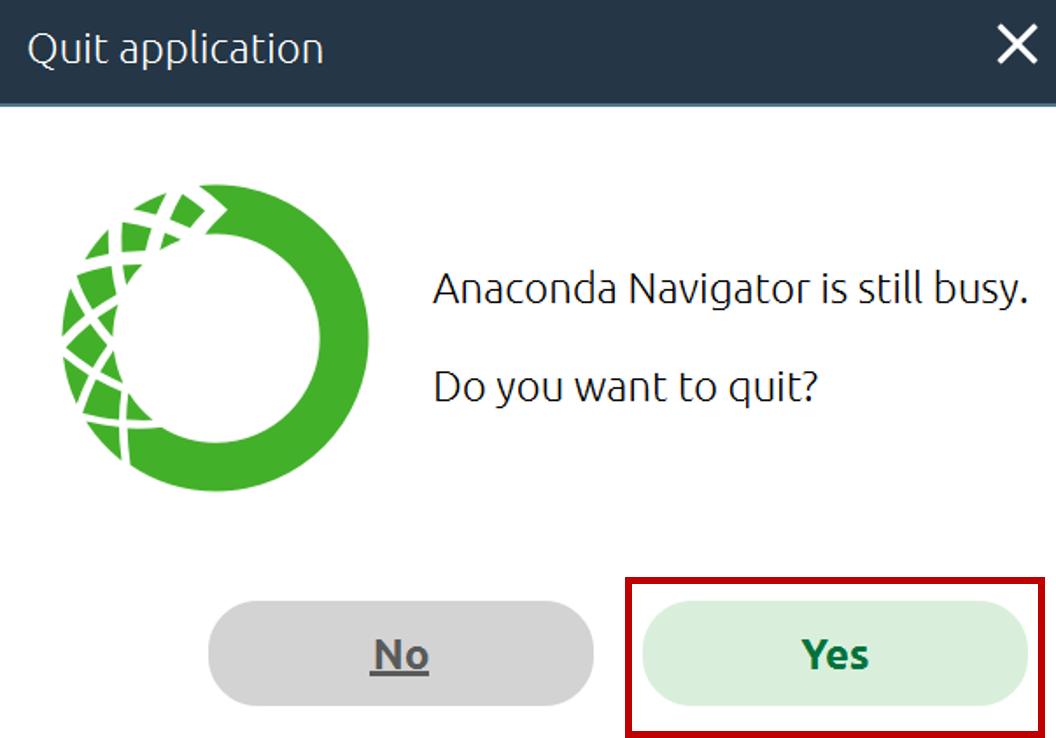
- If prompted to quit Anaconda, click Yes.
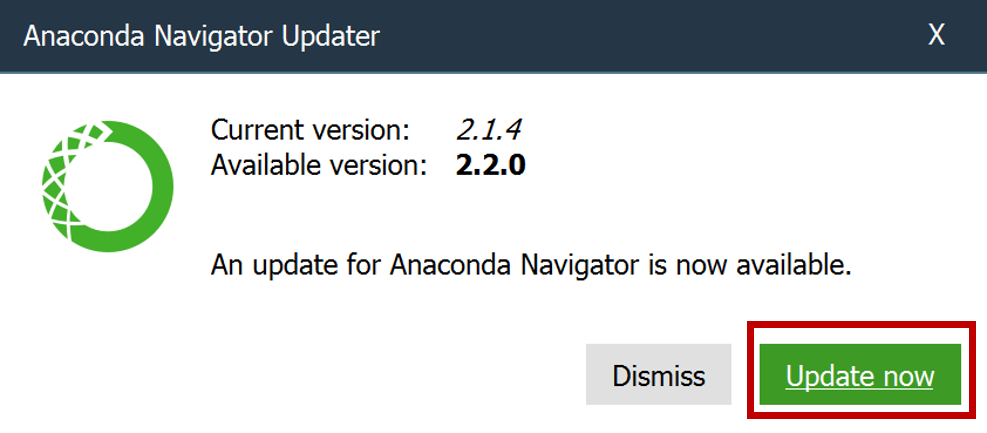
- If prompted again to update, click Update now.
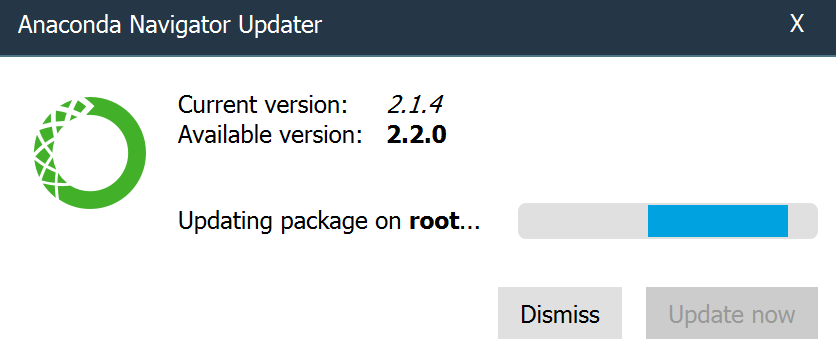
- When the update is complete, click Launch Navigator.
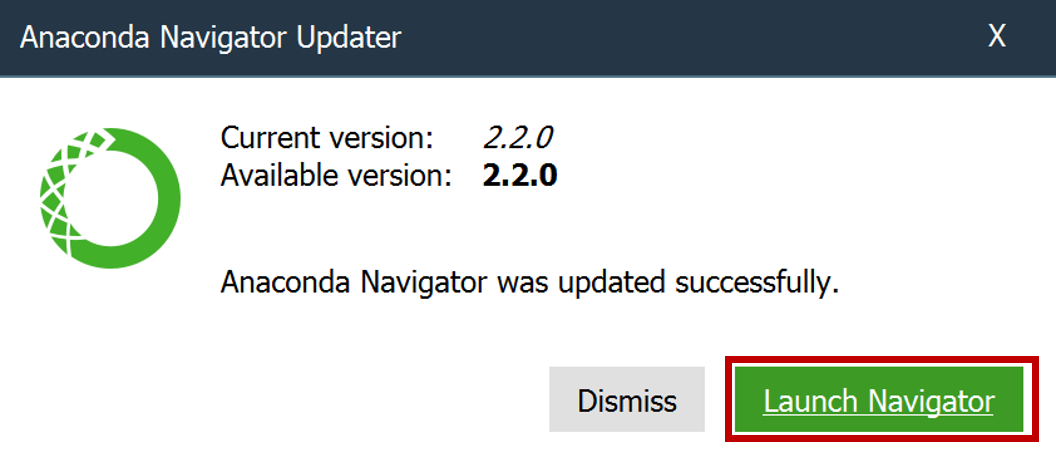
- Launch Anaconda Navigator and click on Environments.

- Download the environment you need:
- Scanpy (to run ComBat, Leiden, UMAP and densMAP)
- Cytonormpy
- hdbscan
- Ivis
- openTSNE
- PacMAP
- PARC (to run PARC, UMAP and densMAP)
- Phate
- Trimap
- UMAP and densMAP
- [FCS Express legacy environment - Y2022]
- In Anaconda > Environments, click Import.
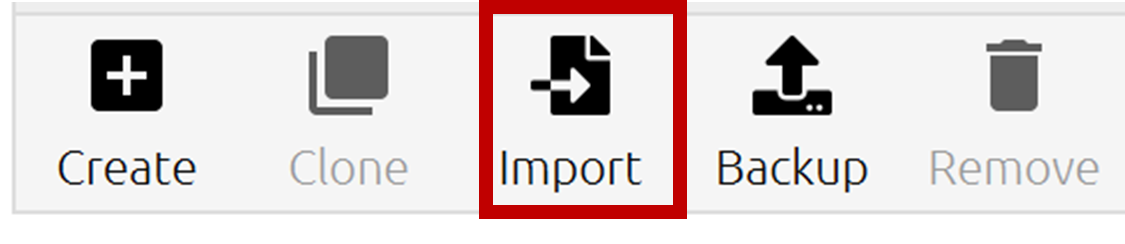
- Select Local drive and click on the File Folder icon next to it
- Find the *.yml environment file that you downloaded from the website. It may be in your computer’s "Downloads" folder.
- Click Open.
- Click Import.
- The environment has now been loaded into your Anaconda Navigator.
If the Python directory you wish to use in FCS Express is the Default, Registered Python on your computer, you may skip this step. The Default, Registered Python can be found in Windows RegistryEditor > Computer\HKEY_CURRENT_USER\Software\Python\PythonCore\3.X\InstallPath
- Launch FCS Express version 7.26 or later.
- Open a New Layout
- Click File > Options.
- In the left pane, click Files/Directories
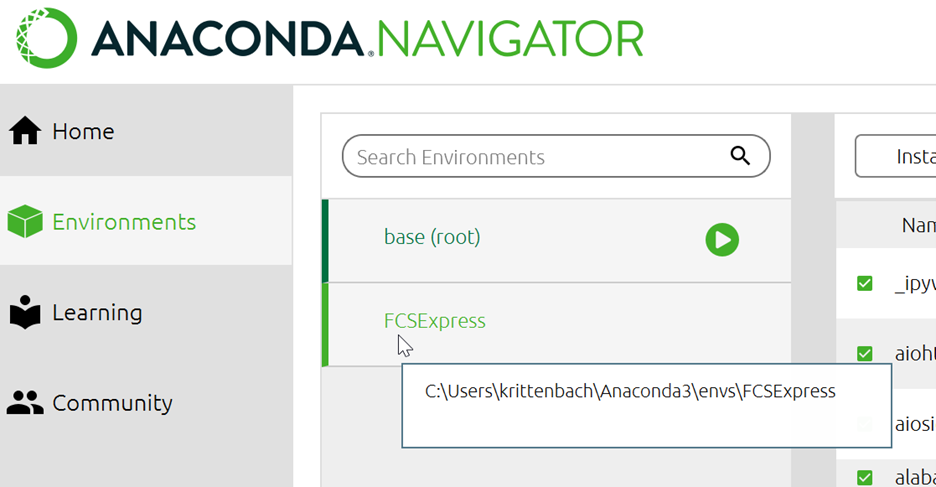
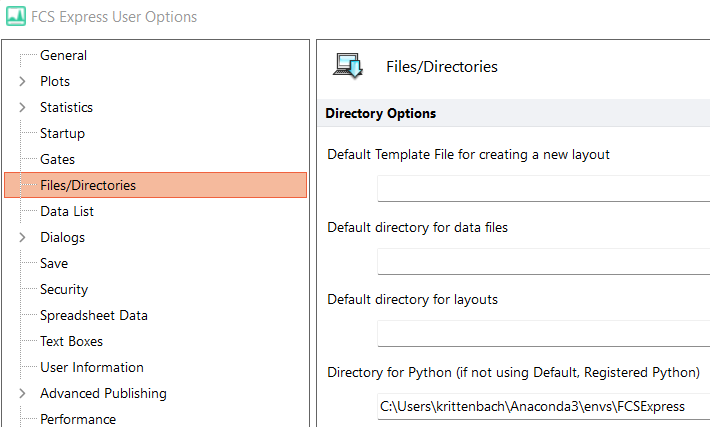
- Under Directory for Python (if not using Default, Registered Python), enter the path where the pythonXXX.dll file is located for the environment you wish to use. If you have installed Python via Anaconda, and wish to use any of the environment above, enter the path to that environment. Said path is found by hovering over the Anaconda environment (e.g. C:\Users\krittenbach\Anaconda3\envs\FCSExpress in the screenshot below)

-
-
- If desired, the Python directory (for an Anaconda environment) can alternatively be found and copied by the following steps:
- Click on the desired environment name in the Anaconda Navigator.
- Click the green "play" button next to the environment name.
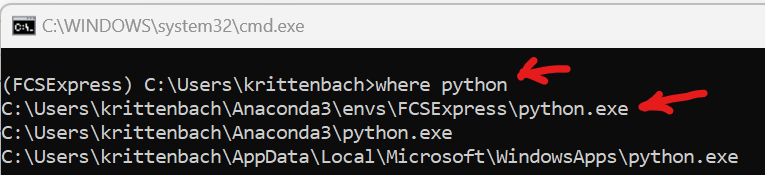
- Click Open Terminal
- Enter the following text: where python and press the Enter key
- Copy the path that contains your environment (i.e. C:\Users\krittenbach\Anaconda3\envs\FCSExpress)
- If desired, the Python directory (for an Anaconda environment) can alternatively be found and copied by the following steps:
-
-
-
-
- Paste the path into the FCS Express user option (Files/Directories > Directory for Python)
-
-
-
- If you installed Python from python.org, the path is likely similar to: C:\Users\XXXXXXXX\AppData\Local\Programs\Python\PythonXXX
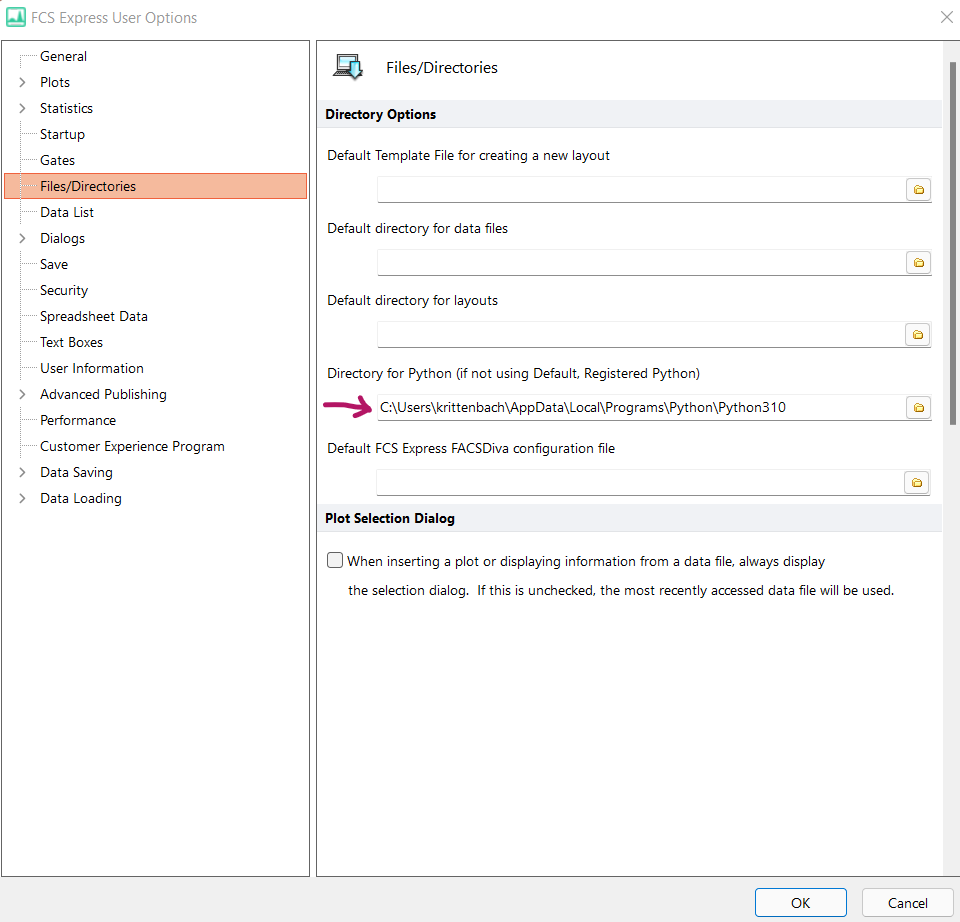
- In FCS Express, click OK to apply the new user option.
- You can now try using the example scripts (provided on this page) in FCS Express.
- Once a Pipeline is created, the Python transformation pipeline step can be added from the + > Miscellaneous category.
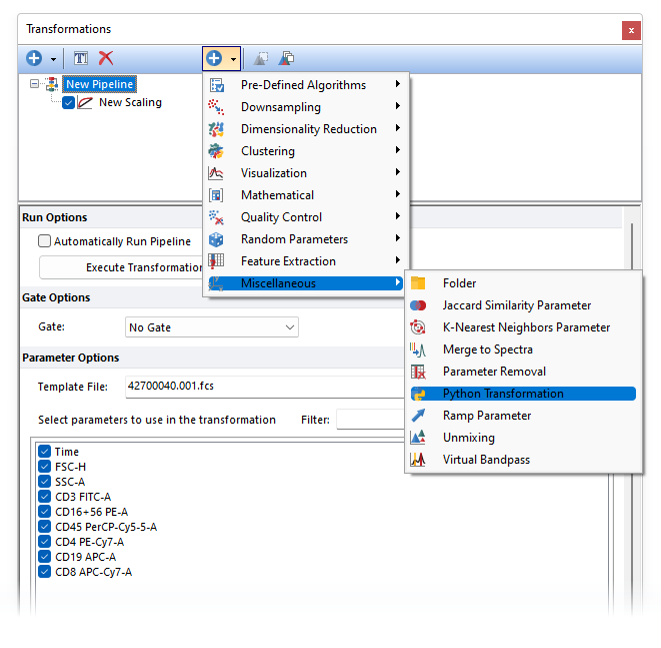
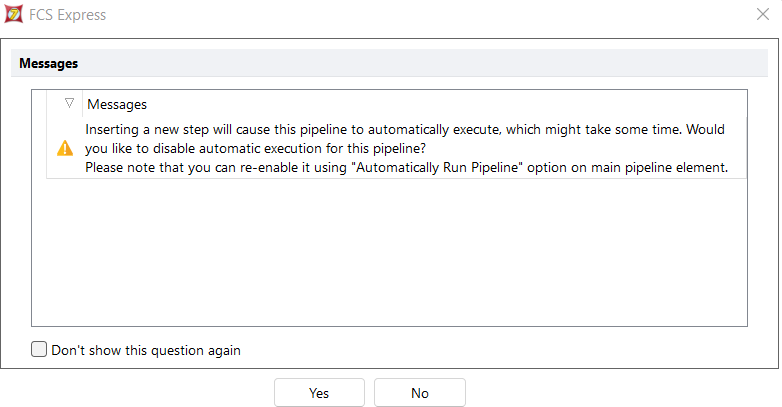
- If the pipeline is already applied to a plot, you may get the warning below asking whether you want to disable the automatic execution of the pipeline and control the execution by the "Execute Transformation Pipeline" button instead.
- The Python Transformation pipeline steps will be added to the pipeline.
- Select the input parameters of interest.
- From one of the example scripts included on this webpage (below), highlight all of the text beginning with the "#". Press Ctrl+C or right-click>Copy.
- In the Python Script text box, right right-click and choose Clear Script to clear the example script .
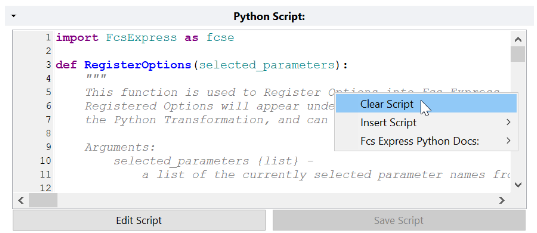
- Ensure the cursor is in the left-most position of the first line. To confirm this, press the Home button on your keyboard.
- In FCS Express, click inside the Python Script text box and press Ctrl+V to paste the script. Note: Right-Click > Paste is not available.
- Click Save Script.
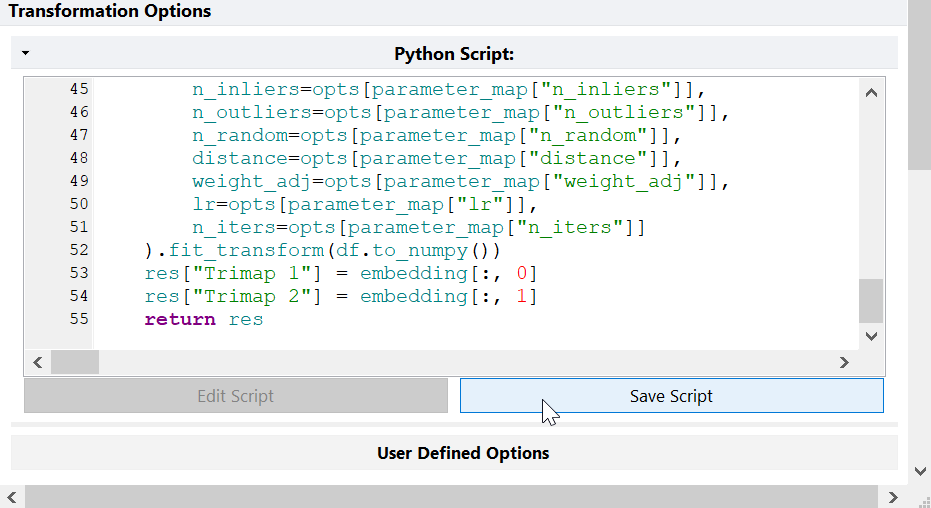
- The User Defined Options section of the Python Transformation step will then be populated based on the script code.
- Drag the Pipeline from the Transformations window onto the plot in the layout.

- If you do not have "Automatically Run Pipeline" checked, press Execute Transformation Pipeline in the pipeline root step.
- Running the pipeline may take a few seconds or several minutes, depending on the file size, chosen script and computer power.
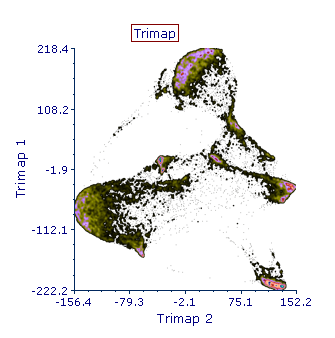
- Once the transformation is applied to a plot and the run is completed, the output parameters generated by the pipeline will be accessible as any other classical parameters by clicking on the X and Y axis title of the plot of interest (e.g. Trimap 1 and Trimap 2 in the screenshot below).
Note: When the script is done running, the title of your plot will say "Pipeline transformed". In case your plot title does not contain the filename, you can check if the transformation is applied in the Overlay formatting dialog for that plot.
Some example scripts are presented below. Please see our full documentation on using the Python pipeline step in the FCS Express manual for more information on creating and implementing scripts.
In this example script, the Python Transformation pipeline step is defined to run CytonormPy (https://cytonormpy.readthedocs.io/en/latest/index.html), which is the Python port of Cytonorm (Van Gassen S, Gaudilliere B, Angst MS, Saeys Y, Aghaeepour N. CytoNorm: A Normalization Algorithm for Cytometry Data. Cytometry A. 2020 Mar;97(3):268-278. doi: 10.1002/cyto.a.23904. Epub 2019 Oct 21. PMID: 31633883; PMCID: PMC7078957.)
Note: To run this script:
- Complete Steps 1 to 5 above.
- Create the metadata.CSV file with the following mandatory columns:
- file_name. This column must include sample filenames. Order should match the order in which files are merged into FCS Express (i.e. the order in the Classification Identifier parameter).
- reference. This column must be used to label each sample as either "other" or "ref" (without quotes).
- batch. This column must be used to identify the sample batch using integer numbers.
- No additional columns are allowed
An example of metadata.CSV file is showed below:
| file_name | reference | batch |
| Gates_PTLG021_IFNa_LPS_Control_1.fcs | other | 1 |
| Gates_PTLG021_IFNa_LPS_Control_2.fcs | other | 1 |
| Gates_PTLG021_Unstim_Control_1.fcs | ref | 1 |
| Gates_PTLG021_Unstim_Control_2.fcs | other | 1 |
| Gates_PTLG025_IFNa_LPS_Control_1.fcs | other | 2 |
| Gates_PTLG025_IFNa_LPS_Control_2.fcs | other | 2 |
| Gates_PTLG025_Unstim_Control_1.fcs | ref | 2 |
| Gates_PTLG025_Unstim_Control_2.fcs | other | 2 |
| Gates_PTLG026_IFNa_LPS_Control_1.fcs | other | 3 |
| Gates_PTLG026_IFNa_LPS_Control_2.fcs | other | 3 |
| Gates_PTLG026_Unstim_Control_1.fcs | ref | 3 |
| Gates_PTLG026_Unstim_Control_2.fcs | other | 3 |
| Gates_PTLG027_IFNa_LPS_Control_1.fcs | other | 4 |
| Gates_PTLG027_IFNa_LPS_Control_2.fcs | other | 4 |
| Gates_PTLG027_Unstim_Control_1.fcs | ref | 4 |
| Gates_PTLG027_Unstim_Control_2.fcs | other | 4 |
| Gates_PTLG028_IFNa_LPS_Control_1.fcs | other | 5 |
| Gates_PTLG028_IFNa_LPS_Control_2.fcs | other | 5 |
| Gates_PTLG028_Unstim_Control_1.fcs | ref | 5 |
| Gates_PTLG028_Unstim_Control_2.fcs | other | 5 |
| ... | ... | ... |
# This script allows you to run CytoNormPy as a Python Transformation pipeline step in FCS Express 7
# Detailed info on CytoNormPy in Python can be found at https://cytonormpy.readthedocs.io/en/latest/index.html
from FcsExpress import *
import importlib
import warnings
missing_module_error = "Current Python distribution does not contain {0}. " \
"To test this script please install {0}. " \
"For more information please visit ({1})"
# Required modules and their aliases
module_aliases = {
"pandas": "pd",
"numpy": "np",
"anndata": "ad",
"cytonormpy": "cnp"
}
# Websites to obtain required modules
module_websites = {
"pandas": "https://pandas.pydata.org/pandas-docs/stable/getting_started/install.html",
"numpy": "https://numpy.org/",
"anndata": "https://anndata.readthedocs.io/en/stable/",
"cytonormpy": "https://cytonormpy.readthedocs.io/en/latest/index.html"
}
# Import required modules
for module_name, module_alias in module_aliases.items():
try:
globals()[module_alias] = importlib.import_module(module_name)
except ModuleNotFoundError:
raise RuntimeError(missing_module_error.format(module_name, module_websites[module_name]))
# Define variables for options to avoid mistyping and improve readability
num_flowsom_clusters = "Number of FlowSOM clusters"
cv_threshold = "Cluster cv threshold"
num_jobs = "Number of jobs"
metadata_path = "Metadata Path"
output_param_suffix = "Suffix for output parameters"
show_warnings = "Show Warnings from cytonormpy"
# Some helper parameters
valid_identifier = "Classification Identifier"
invalid_identifier = "File Identifier"
# Define the metadata structure that is required for Execute method to run properly
metadata_schema = {
'required_columns': ['file_name', 'reference', 'batch'],
'optional_columns': ['sample_id'],
# Strings are stored as object type
'column_types': {'file_name': 'object', 'reference': 'object', 'batch': 'int64'},
'metadata_constraints': {
'reference': {'allowed_values': ['ref', 'other']},
'batch': {'min_value': 1}
}
}
def RegisterOptions(params):
RegisterIntegerOption(num_flowsom_clusters, 10)
RegisterIntegerOption(cv_threshold, 2)
RegisterIntegerOption(num_jobs, 8)
RegisterStringOption(metadata_path, "\\path\\to\\metadata\\csv\\file")
RegisterStringOption(output_param_suffix, "CytonormPy Normalized")
RegisterBooleanOption(show_warnings, True)
def RegisterParameters(opts, params):
for cur_param in params:
if cur_param not in [valid_identifier, invalid_identifier]:
RegisterParameter(f"{cur_param} {opts[output_param_suffix]}")
def Execute(opts, data, result):
df = pd.DataFrame(data)
# Remove File Identifier if it exists, which should not be used as its a numeric value and
# Classification Identifier can more easily be used to identify files since its a textual value
# If File Identifier has been accidentally selected as an input parameter it will be removed
if invalid_identifier in df.columns:
df = df.drop(invalid_identifier, axis = 1)
# Saving file IDs from the "Classification Identifier" parameter, then remove it from df
# File IDs will be used to count how many events each file contains, which will
# be used to create the obs_df object by repeating metadata in the right order
# Using .values on the dataframe to get the file IDs into a numpy array to pass to numpy
if valid_identifier in df.columns:
classification_identifier = df[valid_identifier].values
events = df.drop(valid_identifier, axis = 1)
else:
raise Exception(f"The {valid_identifier} parameter must be selected")
# Load metadata from user specified path
metadata = pd.read_csv(opts[metadata_path])
# Make header lowercase so that validation can be case insensitive
metadata.columns = metadata.columns.str.lower()
# Add to metadata_schema now that more information is available
num_files = len(InputClassificationLabels[valid_identifier])
current_metadata_schema = metadata_schema
current_metadata_schema['num_rows'] = num_files
current_metadata_schema['metadata_constraints']['batch']['max_value'] = num_files
current_metadata_schema['metadata_constraints']['file_name'] = {
'allowed_values': InputClassificationLabels[valid_identifier]
}
# If the metadata contains a sample_ID column we will validate it as well using
# the same value constraints being used for the batch column
if 'sample_id' in metadata.columns:
current_metadata_schema['column_types']['sample_id'] = 'int64'
current_metadata_schema['metadata_constraints']['sample_id'] = current_metadata_schema['metadata_constraints']['batch']
metadata_validation_issues = validate_metadata(metadata, current_metadata_schema, opts)
if metadata_validation_issues:
raise Exception(f"Metadata does not have the correct schema. Issues with metadata found:\n" \
f"{chr(10).join(metadata_validation_issues)}")
#Count how many events each file contains
num_events_per_file = np.unique(classification_identifier, return_counts = True)[1]
# Creating the obs_df object containing metadata (i.e. reference and batch labels) for each data point
# In FCS Express, Classification Identifier values are always ordered (e.g. first all events with classifier equal to 0,
# then all events with classifier equal to 1,...) so we can use "repeat()" to repeat each row of the metadata table,
# the number of events times for its corresponding file
# The match is done using ".index", which start from 0 in the metadata table and thus match the numeration in the
# classification_identifier (which also starts at 0). Note that this can only be done if files are sorted in the same way
# in the input data frame (i.e. from FCS Express) and in the metadata csv file
metadata_order_valid = InputClassificationLabels[valid_identifier] == list(metadata["file_name"])
# If metadata order is incorrect try to sort the metadata table by file_name and see if it matches the Classification Identifier
if not metadata_order_valid:
metadata = metadata.sort_values(by = ["file_name"])
metadata_order_valid = InputClassificationLabels[valid_identifier] == list(metadata["file_name"])
if metadata_order_valid:
print("Metadata table has been sorted alphabetically by the file_name column")
if metadata_order_valid:
obs_df = metadata.loc[metadata.index.repeat(num_events_per_file)].reset_index(drop = True)
else:
raise Exception("Samples must be sorted the same way in the merged data and in the metadata csv, and file names must match.")
# Create the var_df dataframe containing metadata for variables
var_df = pd.DataFrame({"channels": events.columns})
# Create the annotation data
# the annotation data is expecting the row indexs to be strings
# so we can convert the index first
obs_df.index = obs_df.index.astype(str)
var_df.index = var_df.index.astype(str)
adata = ad.AnnData(
obs = obs_df,
var = var_df,
layers = {"compensated": events}
)
with warnings.catch_warnings():
if not opts[show_warnings]:
warnings.simplefilter("ignore")
else:
warnings.formatwarning = fcse_warning_format
# Create the Cytonorm object using FlowSOM as clusterer
cn = cnp.CytoNorm()
fs = cnp.FlowSOM(n_clusters = opts[num_flowsom_clusters])
cn.add_clusterer(fs)
cn.run_anndata_setup(adata, layer = "compensated", key_added = "normalized")
cn.run_clustering(cluster_cv_threshold = opts[cv_threshold])
cn.calculate_quantiles()
cn.calculate_splines(goal = "batch_mean")
cn.normalize_data(n_jobs = opts[num_jobs])
for index, new_param in enumerate(result):
result[new_param] = adata.layers["normalized"][:, index]
# Return the result object that will be re-imported back into FCS Express
return result
# Define our metadata validation function so that issues with metadata are presented up front
# rather than via an error that occurs when using the metadata which will be harder to parse
def validate_metadata(metadata_df, valid_metadata_schema, opts):
validation_issues = []
if metadata_df.empty:
validation_issues.append(f"Metadata contains no data. Please check file located at: ({opts[metadata_path]})")
# We can skip further validation as there is nothing to validate
return validation_issues
if 'required_columns' in valid_metadata_schema:
required_cols = set(valid_metadata_schema['required_columns'])
actual_cols = set(metadata_df.columns)
missing_cols = required_cols - actual_cols
extra_cols = actual_cols - required_cols
if 'optional_columns' in valid_metadata_schema:
extra_cols = extra_cols - set(valid_metadata_schema['optional_columns'])
if missing_cols:
validation_issues.append(f"Metadata is missing required columns: {sorted(missing_cols)}")
if extra_cols:
validation_issues.append(f"Metadata contains unexpected columns: {sorted(extra_cols)}")
if missing_cols or extra_cols:
validation_issues.append(f"Columns required in Metadata: {valid_metadata_schema['required_columns']}")
validation_issues.append(f"Columns found in Metadata: {list(metadata_df.columns)}")
# We can skip further validation as the columns being correct is required
return validation_issues
if 'column_types' in valid_metadata_schema:
for col, expected_type in valid_metadata_schema['column_types'].items():
if str(metadata_df[col].dtype) != expected_type:
# Rename type for error message as object is not clear
if expected_type == 'object':
expected_type = 'string'
validation_issues.append(f"Column '{col}' has type '{str(metadata_df[col].dtype)}', expected '{expected_type}'")
elif expected_type == 'object':
for index, row_val in enumerate(metadata_df[col]):
if type(row_val) != str:
validation_issues.append(f"Column '{col}' is expected to contain only strings. " \
f"However, row #{index + 1} has a type of '{type(row_val)}'")
if 'num_rows' in valid_metadata_schema and len(metadata_df) != valid_metadata_schema['num_rows']:
validation_issues.append(f"Metadata contains the incorrect number of rows. " \
f"There should be one row per file ID found in 'Classification Identifier'. " \
f"Metadata contains {len(metadata_df)} rows. " \
f"Number of file IDs = {valid_metadata_schema['num_rows']}")
if 'metadata_constraints' in valid_metadata_schema:
for col, constraints in valid_metadata_schema['metadata_constraints'].items():
if 'allowed_values' in constraints:
allowed_values = set(constraints['allowed_values'])
invalid_values = set(metadata_df[col]) - allowed_values
if invalid_values:
validation_issues.append(f"Column '{col}' has invalid values: {sorted(invalid_values)}. " \
f"Allowed values: {sorted(allowed_values)}")
if 'min_value' in constraints:
if metadata_df[col].min() < constraints['min_value']: validation_issues.append(f"Column '{col}' has values less than allowed minimum: {constraints['min_value']}. " \ f"Minimum found: {metadata_df[col].min()}") if 'max_value' in constraints: if metadata_df[col].max() > constraints['max_value']:
validation_issues.append(f"Column '{col}' has values greater than allowed maximum: {constraints['max_value']}. " \
f"Maximum found: {metadata_df[col].max()}")
return validation_issues
# Create a custom warning format since the location of cytonormpy on disk is not important nor is the line number
# inside cytonormpy that is producing the warning. Instead just the warning message will be printed to keep things simple
def fcse_warning_format(msg, *args, **kwargs):
return 'Cytonormpy Warning: ' + str(msg) + '\n'
In this example script, the Python Transformation pipeline step is defined to run HDBSCAN (Campello R.J.G.B., Moulavi D., Sander J. (2013) Density-Based Clustering Based on Hierarchical Density Estimates. In: Pei J., Tseng V.S., Cao L., Motoda H., Xu G. (eds) Advances in Knowledge Discovery and Data Mining. PAKDD 2013. Lecture Notes in Computer Science, vol 7819. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-37456-2_14).
HDBSCAN can be useful to identify clusters on UMAP plots. When HDBSCA is used for this purpose, creating a UMAP map with dense clusters (i.e. keeping the UMAP "Min Low Dim Distance" value low) will help HDBSCAN clustering.
More information on DBSCAN for Python can be found here.
Note #1: to run this script, please first complete Steps 1 to 5 above.
Note #2: If noisy data points are detected by HDBSCA, they will be grouped under "HDBSCAN Cluster Assignment" equal to "1" in FCS Express.
Note #3: with large dataset, HDBSCAN might return a "Could not find -c" error. If that happens, setting core_dist_n_jobs to 1 might solve the issue.
#HDBSCAN
from FcsExpress import *
try:
from pandas import DataFrame
except ModuleNotFoundError:
raise RuntimeError("Current Python distribution does not contain Pandas. " +
"To test this script please install pandas: " +
"(https://pandas.pydata.org/pandas-docs/stable/getting_started/install.html)")
try:
import hdbscan
except ModuleNotFoundError:
raise RuntimeError("Current Python distribution does not contain hdbscan. " +
"To test this script please install hdbscan: " +
"(https://pypi.org/project/hdbscan/)")
from collections import Counter
def RegisterOptions(params):
RegisterIntegerOption("min_cluster_size",5)
RegisterStringOption("min_samples","None")
RegisterIntegerOption("core_dist_n_jobs",4)
def RegisterParameters(opts, params):
RegisterClassification("HDBSCAN Cluster Assignments")
RegisterParameter("HDBSCAN Outlier scores")
def Execute(opts, data, res):
df = DataFrame(data)
#See if doing default min_samples
if opts["min_samples"] == "None":
min_samples = None # Setting n_neighbors to "None" leads to a default choice
elif opts["min_samples"].isnumeric():
min_samples = int(opts["min_samples"])
else:
raise Exception("The min_samples option only accept None or integer values")
print("Running HDBSCAN on {} data points, with min_cluster_size={}, min_samples={} and core_dist_n_jobs={}".format(NumberDataPoints,opts["min_cluster_size"],min_samples,opts["core_dist_n_jobs"]))
clusterer = hdbscan.HDBSCAN(min_cluster_size=opts["min_cluster_size"],
min_samples=min_samples,
core_dist_n_jobs=opts["core_dist_n_jobs"])
cluster_labels = clusterer.fit_predict(df)
outlier_scores=clusterer.outlier_scores_
#Noisy data points are labelled by HDBScan with cluster assignemnt of -1.
#FCS Express requires cluster assignment values starting from 0.
#So, in case noisy data points are present, cluster labels are increased by 1
if min(cluster_labels)==-1:
noisy=Counter(cluster_labels)[-1] #number of noisy data points
print(noisy, "noisy datapoints have been detected" +
" and will be grouped under HDBSCAN Cluster Assignment equal to 1 in FCS Express")
cluster_labels=cluster_labels+1 # making cluster assignment values starting at 0.
res["HDBSCAN Cluster Assignments"] = cluster_labels
res["HDBSCAN Outlier scores"] = outlier_scores
return res
In this example script, the Python Transformation pipeline step is defined to run Ivis (Szubert, B., Cole, J.E., Monaco, C. et al. Structure-preserving visualisation of high dimensional single-cell datasets. Sci Rep 9,8914 (2019). https://doi.org/10.1038/s41598-019-45301-0). More information on Ivis for Python can be found here and here.
Note: to run this script please first complete Steps 1 to 5 above.
#This script allows you to run Ivis as a Python Transformation pipeline step in FCS Express 7 #Detailed info on Ivis in Python can be found at https://bering-ivis.readthedocs.io/en/latest/python_package.html from FcsExpress import * try: from pandas import DataFrame except ModuleNotFoundError: raise RuntimeError("Current Python distribution does not contain Pandas. " + "To test this script please install pandas: " + "(https://pandas.pydata.org/pandas-docs/stable/getting_started/install.html)") try: from ivis import Ivis except ModuleNotFoundError: raise RuntimeError("Current Python distribution does not contain ivis. " + "To test this script please install ivis: " + "(https://github.com/beringresearch/ivis)") try: from scipy import sparse except ModuleNotFoundError: raise RuntimeError("Current Python distribution does not contain scipy. " + "To test this script please install scypy: " + "(https://scipy.org/)") def RegisterOptions(params): RegisterIntegerOption("Embedding Dimensions",2) RegisterIntegerOption("Number of Nearest Neighbours",15) RegisterIntegerOption("Number of epochs without progress",10) RegisterStringOption("Type of input", "numpy array") RegisterStringOption("Keras Model", "auto") #The Number of Nearest Neighbours , the Number of epochs without progress #and the Keras Model are tunable parameters that should be selected #on the basis of dataset size and complexity. #Please refer to https://bering-ivis.readthedocs.io/en/latest/hyperparameters.html def RegisterParameters(opts, params): for dim in range(opts["Embedding Dimensions"]): RegisterParameter("Ivis {}".format(dim + 1)) def Execute(opts, data, res): df = DataFrame(data) #Converting the input data to either numpy array or spare matrix based on the user selection if opts["Type of input"]=="numpy array": X=df.to_numpy() elif opts["Type of input"]=="sparse matrix": X=sparse.csr_matrix(df) else: raise Exception("The Type of input option only accept the following values: numpy array, sparse matrix") # Determine model. For more details, see: # https://bering-ivis.readthedocs.io/en/latest/hyperparameters.html models = ["maaten","szubert","hinton"] if opts["Keras Model"]=="auto": if NumberDataPoints > 500000: keras_model = "szubert" else: keras_model = "maaten" elif opts["Keras Model"] in models: keras_model = opts["Keras Model"] else: raise Exception("The Model option only accept the following values: auto, maaten, szubert, hinton") print("Running Ivis with keras model {} on {} data points, using {} as input data type".format(keras_model, NumberDataPoints,opts["Type of input"])) # Set ivis parameters model = Ivis(embedding_dims = opts["Embedding Dimensions"], k = opts["Number of Nearest Neighbours"], n_epochs_without_progress = opts["Number of epochs without progress"], model = keras_model ) embeddings = model.fit_transform(X) # Generate embeddings #Loop over dimensions and set results for dim in range(opts["Embedding Dimensions"]): res["Ivis {}".format(dim + 1)] = embeddings[:, dim] return res #return the res object that will be re-imported into FCS Express
In this example script, the Python Transformation pipeline step is defined to run openTSNE (openTSNE: A Modular Python Library for t-SNE Dimensionality Reduction and Embedding. Poličar, P.G., Stražar, M. and Zupan, B. Journal of Statistical Software. 109, 3 (May 2024), 1–30. DOI:https://doi.org/10.18637/jss.v109.i03).
More details on openTSNE can be found here.
Note: to run this script please first complete Steps 1 to 5 above.
#This script allows you to run openTSNE as a Python Transformation pipeline step in FCS Express 7 #Detailed info on openTSNE in Python can be found at https://opentsne.readthedocs.io/en/latest/index.html from FcsExpress import * try: from pandas import DataFrame except ModuleNotFoundError: raise RuntimeError("Current Python distribution does not contain Pandas. " + "To test this script please install pandas: " + "(https://pandas.pydata.org/pandas-docs/stable/getting_started/install.html)") try: from openTSNE import TSNE except ModuleNotFoundError: raise RuntimeError("Current Python distribution does not contain OpenTSNE. " + "To test this script please install openTSNE: " + "(https://opentsne.readthedocs.io/en/latest/installation.html)") def RegisterOptions(params): RegisterIntegerOption("Number Iterations", 500) RegisterIntegerOption("Perplexity", 30) RegisterFloatOption("Burnes-Hut Theta", 0.5) RegisterStringOption("Learning Rate", "auto") RegisterStringOption("Metric", "euclidean") RegisterIntegerOption("Random State", 42) RegisterStringOption("Initialization","pca") def RegisterParameters(opts, params): RegisterParameter("openTSNE 1") RegisterParameter("openTSNE 2") def Execute(opts, data, res): df = DataFrame(data) # See if doing auto learning if opts["Learning Rate"] == "auto": l_rate = 'auto' # Auto learning will be used elif opts["Learning Rate"].isnumeric(): l_rate = float(opts["Learning Rate"]) else: raise Exception("The Learning Rate option only accept auto or floating values") #Check initialization Initializations=["pca","spectral","random"] if opts["Initialization"] not in Initializations: raise Exception("The Initialization option only accept the following values: pca, spectral, random") #Run TSNE embedding = TSNE( theta=opts["Burnes-Hut Theta"], perplexity=opts["Perplexity"], n_iter=opts["Number Iterations"], learning_rate=l_rate, metric=opts["Metric"], random_state=opts["Random State"], initialization=opts["Initialization"] ).fit(df.to_numpy()) print("Running TSNE with {} Initialization on {} data points, with Number of Iterations = {}, Perplexity = {}, Theta = {}, Learning Rate = {}, Metric = {}, Random State = {}".format(opts["Initialization"], NumberDataPoints, opts["Number Iterations"], opts["Perplexity"],opts["Burnes-Hut Theta"],l_rate,opts["Metric"],opts["Random State"])) res["openTSNE 1"] = embedding[:, 0] res["openTSNE 2"] = embedding[:, 1] return res
In this example script, the Python Transformation pipeline step is defined to run PaCMAP (Wang and Huang, at al., Understanding How Dimension Reduction Tools Work: An Empirical Approach to Deciphering t-SNE, UMAP, TriMap, and PaCMAP for Data Visualization; Journal of Machine Learning Research 22 (2021) 1-73). More information on PaCMAP for Python can be found here.
Note: to run this script, please first complete Steps 1 to 5 above.
#This script allows you to run PaCMAP as a Python Transformation pipeline step in FCS Express 7 #Detailed info on PaCMAP in Python can be found at https://github.com/YingfanWang/PaCMAP from FcsExpress import * try: from pandas import DataFrame except ModuleNotFoundError: raise RuntimeError("Current Python distribution does not contain Pandas. " + "To test this script please install pandas: " + "(https://pandas.pydata.org/pandas-docs/stable/getting_started/install.html)") try: import pacmap except ModuleNotFoundError: raise RuntimeError("Current Python distribution does not contain Pacmap. " + "To test this script please install pacmap: " + "(https://github.com/YingfanWang/PaCMAP)") def RegisterOptions(params): RegisterIntegerOption("Number of dimensions", 2) RegisterStringOption("Number of neighbors", "None") RegisterFloatOption("MN_ratio", 0.5) RegisterFloatOption("FP_ratio", 2.0) RegisterStringOption("Initialization","pca") RegisterIntegerOption("Random seed", 4) RegisterIntegerOption("Number of iterations", 450) def RegisterParameters(opts, params): for dim in range(opts["Number of dimensions"]): RegisterParameter("PaCMAP {}".format(dim + 1)) def Execute(opts, data, res): #Reading data df = DataFrame(data) X = df.to_numpy() #See if doing default n_neighbors if opts["Number of neighbors"] == "None": n_neighbors = None # Setting n_neighbors to "None" leads to a default choice elif opts["Number of neighbors"].isnumeric(): n_neighbors = int(opts["Number of neighbors"]) else: raise Exception("The Number of neighbors option only accept None or integer values") #Check initialization Initializations=["pca","random"] if opts["Initialization"] not in Initializations: raise Exception("The Initialization option only accept the following values: pca, random") #Initializing the pacmap instance embedding = pacmap.PaCMAP( n_components=opts["Number of dimensions"], n_neighbors=n_neighbors, MN_ratio=opts["MN_ratio"], FP_ratio=opts["FP_ratio"], random_state=opts["Random seed"], num_iters=opts["Number of iterations"], verbose=True) #Running PaCMAP X_transformed = embedding.fit_transform(X, init=opts["Initialization"]) print("Running PaCMAP with {} Initialization on {} data points, with Number of Neighbors set to {}".format(opts["Initialization"], NumberDataPoints, n_neighbors)) #Loop over dimensions and set results for dim in range(opts["Number of dimensions"]): res["PaCMAP {}".format(dim + 1)] = X_transformed[:, dim] return res
In this example script, the Python Transformation pipeline step is defined to run PARC (PARC: ultrafast and accurate clustering of phenotypic data of millions of single cells; Shobana V Stassen; Bioinformatics, Volume 36, Issue 9, 1 May 2020). More details on parc can be found here.
Note: to run this script, please first complete Steps 1 to 5 above.
#This script allows you to run PARC as a Python Transformation pipeline step in FCS Express 7)
#Detailed info on PARC in Python can be found at https://github.com/ShobiStassen/PARC
from FcsExpress import *
try:
from pandas import DataFrame
except ModuleNotFoundError:
raise RuntimeError("Current Python distribution does not contain Pandas. " +
"To test this script please install pandas: " +
"(https://pandas.pydata.org/pandas-docs/stable/getting_started/install.html)")
try:
import parc
except ModuleNotFoundError:
raise RuntimeError("Current Python distribution does not contain parc. " +
"To test this script please install parc: " +
"(https://github.com/ShobiStassen/PARC)")
def RegisterOptions(params):
RegisterIntegerOption("dist_std_local",2)
RegisterStringOption("jac_std_global","median")
RegisterIntegerOption("resolution_parameter",1)
def RegisterParameters(opts, params):
RegisterClassification("Parc Cluster Assignments")
def Execute(opts, data, res):
df = DataFrame(data)
#Function to check whether an object is float
def isfloat(num):
try:
float(num)
return True
except ValueError:
return False
#checking whether opts["jac_std_global"] is set to median or whether it is float
if opts["jac_std_global"]=="median":
jac_std_global="median"
elif isfloat(opts["jac_std_global"])==True:
jac_std_global=float(opts["jac_std_global"])
else:
raise Exception("The jac_std_global only accept median or numerical values")
print("Running PARC with dist_std_local {}, jac_std_global {}, resolution_parameter {}, on {} data points".format(opts["dist_std_local"],jac_std_global,opts["resolution_parameter"],NumberDataPoints))
parc_obj = parc.PARC(df.to_numpy(),dist_std_local=opts["dist_std_local"],jac_std_global=jac_std_global,resolution_parameter=opts["resolution_parameter"]) #initiate PARC
parc_obj.run_PARC() #run the clustering
res["Parc Cluster Assignments"] = parc_obj.labels #saving the cluster assignment into the res object
return res #return the res object that will be re-imported into FCS Express
In this example script, the Python Transformation pipeline step is defined to run Phate (Moon, K.R., van Dijk, D., Wang, Z. et al. Visualizing structure and transitions in high-dimensional biological data. Nat Biotechnol 37, 1482–1492 (2019). https://doi.org/10.1038/s41587-019-0336-3). More information on Phate for Python can be found here.
Note: to run this script, first complete Steps 1 to 5 above.
#This script allows you to run Phate as a Python Transformation pipeline step in FCS Express 7.
#Detailed info on Phate in Python can be found at https://pypi.org/project/phate/
from FcsExpress import *
try:
from pandas import DataFrame
except ModuleNotFoundError:
raise RuntimeError("Current Python distribution does not contain Pandas. " +
"To test this script please install pandas: " +
"(https://pandas.pydata.org/pandas-docs/stable/getting_started/install.html)")
try:
import phate
except ModuleNotFoundError:
raise RuntimeError("Current Python distribution does not contain phate. " +
"To test this script please install phate: " +
"(https://pypi.org/project/phate/)")
parameter_map = {
"knn": "Number Nearest Neighbors",
"n_components": "Number of dimensions"
}
def RegisterOptions(params):
#The full list of available parameters can be found at #https://phate.readthedocs.io/en/stable/api.html
RegisterIntegerOption(parameter_map["knn"], 5)
RegisterIntegerOption(parameter_map["n_components"], 2)
def RegisterParameters(opts, params):
for dim in range(opts[parameter_map["n_components"]]):
RegisterParameter("Phate {}".format(dim + 1))
def Execute(opts, data, res):
df = DataFrame(data)
phate_operator = phate.PHATE(
knn=opts[parameter_map["knn"]],
n_components=opts[parameter_map["n_components"]]
)
df_phate = phate_operator.fit_transform(df)
#Loop over dimensions and set results
for dim in range(opts[parameter_map["n_components"]]):
res["Phate {}".format(dim + 1)] = df_phate[:, dim]
return res
In this example script, the Python Transformation pipeline step is defined to run TriMap (TriMap: Large-scale Dimensionality Reduction Using Triplets; Ehsan Amid, Manfred K. Warmuth; arXiv:1910.00204; manuscript submitted on 1st Oct 2019 but rejected on 20th Dec 2019). More details on trimap can be found here .
Note: to run this script first complete Steps 1 to 5 above.
#This script allow to run TriMap as a Python Transformation pipeline step in FCS Express 7
#Detailed info on TriMap in Python can be found at https://pypi.org/project/trimap/
from FcsExpress import *
try:
from pandas import DataFrame
except ModuleNotFoundError:
raise RuntimeError("Current Python distribution does not contain Pandas. " +
"To test this script please install pandas: " +
"(https://pandas.pydata.org/pandas-docs/stable/getting_started/install.html)")
try:
import trimap
except ModuleNotFoundError:
raise RuntimeError("Current Python distribution does not contain trimap. " +
"To test this script please install trimap: " +
"(https://pypi.org/project/trimap/)")
parameter_map = {
"n_inliers": "Number Nearest Neighbors",
"n_outliers": "Number Outliers",
"n_random": "Number Random Triplets",
"distance": "Distance Measure",
"weight_adj": "Gamma",
"lr": "Learning Rate",
"n_iters": "Number Iterations"
}
def RegisterOptions(params):
RegisterIntegerOption(parameter_map["n_inliers"], 10)
RegisterIntegerOption(parameter_map["n_outliers"], 5)
RegisterIntegerOption(parameter_map["n_random"], 5)
RegisterStringOption(parameter_map["distance"], "euclidean")
RegisterFloatOption(parameter_map["weight_adj"], 500.0)
RegisterFloatOption(parameter_map["lr"], 1000.0)
RegisterIntegerOption(parameter_map["n_iters"], 400)
def RegisterParameters(opts, params):
RegisterParameter("Trimap 1")
RegisterParameter("Trimap 2")
def Execute(opts, data, res):
df = DataFrame(data)
embedding = trimap.TRIMAP(
n_inliers=opts[parameter_map["n_inliers"]],
n_outliers=opts[parameter_map["n_outliers"]],
n_random=opts[parameter_map["n_random"]],
distance=opts[parameter_map["distance"]],
weight_adj=opts[parameter_map["weight_adj"]],
lr=opts[parameter_map["lr"]],
n_iters=opts[parameter_map["n_iters"]]
).fit_transform(df.to_numpy())
res["Trimap 1"] = embedding[:, 0]
res["Trimap 2"] = embedding[:, 1]
return res
#This script allows you to run UMAP as a Python Transformation pipeline step in FCS Express 7
#Detailed info on UMAP in Python can be found at https://pypi.org/project/umap-learn/
from FcsExpress import *
try:
from pandas import DataFrame
except ModuleNotFoundError:
raise RuntimeError("Current Python distribution does not contain Pandas. " +
"To test this script please install pandas: " +
"(https://pandas.pydata.org/pandas-docs/stable/getting_started/install.html)")
try:
import umap
except ModuleNotFoundError:
raise RuntimeError("Current Python distribution does not contain umap. " +
"To test this script please install umap-learn: " +
"(https://pypi.org/project/umap-learn/)")
def RegisterOptions(params):
RegisterIntegerOption("Number of Neighbours",15)
RegisterFloatOption("Minimum Distance",0.1)
RegisterIntegerOption("Number of Dimensions",2)
RegisterStringOption("Metric", "euclidean")
RegisterStringOption("Number of epochs","None")
RegisterBooleanOption("densMAP","False")
RegisterStringOption("Initialization","spectral")
#The options above, are tunable parameters that should be selected
#on the basis of dataset size and complexity.
#Please refer to https://umap-learn.readthedocs.io/en/latest/api.html
def RegisterParameters(opts, params):
for dim in range(opts["Number of Dimensions"]):
RegisterParameter("UMAP-learn {}".format(dim + 1))
def Execute(opts, data, res):
df = DataFrame(data)
allowed_metrics = ["euclidean","manhattan","chebyshev","minkowski","canberra","braycurtis","haversine","mahalanobis","wminkowski","seuclidean","cosine","correlation","hamming","jaccard","dice","russellrao","kulsinski","ll_dirichlet","hellinger","rogerstanimoto","sokalmichener","sokalsneath","yule"]
# For more details on Metrics, see:
# https://umap-learn.readthedocs.io/en/latest/parameters.html#metric
if opts["Metric"] not in allowed_metrics:
raise Exception("The metric specified is not among the accepted metrics")
#Numer of iterations (epochs)
if opts["Number of epochs"]=="None":
epochs=None
elif type(int(opts["Number of epochs"]))==int:
epochs=int(opts["Number of epochs"])
else:
raise Exception("The Number of Epochs only accept None or integer values")
#densMAP
if opts["densMAP"]:
densMAP=True
else:
densMAP=False
#Initialization
allowed_initializations=["spectral","random","pca"]
if opts["Initialization"] not in allowed_initializations:
raise Exception("The initialization specified is not among the accepted initializations.")
print("Running UMAP with Metric {}, # of Neighbors {}, Min Dist {}, # of Dimensions {}, # of Epochs {}, densMAP {}, initialization {} on {} data points".format(opts["Metric"],opts["Number of Neighbours"],opts["Minimum Distance"],opts["Number of Dimensions"],opts["Number of epochs"],opts["densMAP"],opts["Initialization"],NumberDataPoints))
# Set UMAP parameters
fit = umap.UMAP(n_neighbors=opts["Number of Neighbours"],
min_dist = opts["Minimum Distance"],
n_components = opts["Number of Dimensions"],
metric = opts["Metric"],
n_epochs = epochs,
densmap=densMAP,
init=opts["Initialization"]
)
# Generate embeddings
embeddings = fit.fit_transform(df)
#Loop over dimensions and set results
for dim in range(opts["Number of Dimensions"]):
res["UMAP-learn {}".format(dim + 1)] = embeddings[:, dim]
return res #return the res object that will be re-imported into FCS Express
-
Installation
-
Licenses
-
- Can I get more information regarding the Add-Ons that can be purchased with a license?
- Can I lock my template based on an electronic signature?
- Does FCS Express have any features to help meet 21 CFR Part 11 compliance?
- Does FCS Express have Quality Control features?
- Does FCS Express offer Single Sign On capability?
- How do I configure SQL Server to host a database for FCS Express?
- What database options are available when I purchase the Security option?
- What is the difference between the different types of Users that are available with a Security and Logging license?
- What is the difference between the Logging option and System Level Audit Trails?
- What SQL Server permissions are needed?
-
-
- Can I share my USB dongle or countercode license with another user?
- Can I track usage of the internet dongle?
- Can I try out the Internet Dongle before I make a purchase?
- Can the administrator log users out?
- Do you have to be connected to the internet at all times with the Internet dongle?
- How can users be added to an internet dongle license?
- How do I activate my dongle?
- How do I change my internet dongle/site license password?
- How many people can be logged in at the same time?
- How many user accounts can I create?
- If a user left the computer running can the user log themselves out from another computer?
- What are the differences between the internet dongle and network licensing options?
- What happens if I lose my internet connection?
- What happens if the user leaves the computer without logging out?
- What happens to the users login in case of an unexpected interruption? For instance, a software crash, power failure, etc.
- Why am I receiving a message that FCS Express cannot connect to De Novo Software servers?
- Show all articles ( 1 ) Collapse Articles
-
-
- Can I convert my Cytek license from the countercode licensing option to another licensing option?
- How can I claim my license purchased through BD Accuri Cytometers?
- How can I claim my license purchased through BD Biosciences?
- How can I claim my license purchased through Nexcelom Biosciences?
- How can I claim my license purchased through Sysmex-Partec GmbH?
- How can I claim the FCS Express license that came with my Cytek instrument purchase?
-
-
Layouts & Loading Data
-
- Can I customize my Quick Access Toolbar?
- For an existing layout, can I change the way FCS Express remembers parameters?
- How can I exclude histogram overlays from batch processing?
- How do I change my layout from portrait to landscape?
- How do I change the resolution of my exported images?
- How do I re-establish data files that were linked to a layout?
-
- Are Beckman Coulter LMD files unique?
- Can I find a support resource page for the analysis of Cytek data in FCS Express?
- How can I easily create the "filename" column in the "ExtraKeywords Table.csv" file?
- How can I load a Sony MA900 Index Sort file into FCS Express?
- How can I load data from the BD Accuri C6 Flow Cytometer?
- How can I load MACSQuantify files that were exported from MACSQuantify software version 3.0.1?
- How can I quickly reload all of the data files in the data list?
- How do I change the display in my plots from one data file to another data file?
- How do I export .ICE files from Thermo Cellomics HCS Studio?
- How do I tell FCS Express what plate size to use if that information is not included in the data file?
- How do I upload files to the De Novo Software FTP site?
- How do I use BD Accuri CFlow files with Multicycle DNA analysis in FCS Express?
- How do I work with images from the Thermo Scientific Attune CytPix?
- What is the Elapsed Time setting in the Gallios software and how do I convert it to real time?
- Why am I seeing a warning message when loading my Cytek data onto a layout object?
- Why are iterations in my Data List gray?
- Why are there sometimes access violations when I save and load files?
- Why do I get the message that a data file exported from a FACSDiva™ Experiment is invalid?
- Show all articles ( 3 ) Collapse Articles
-
- How are existing quadrants handled when an old layout is opened in version 7.20 and later?
- How can I set quadrants to behave like conventional gates?
- How can I set quadrants to behave like in earlier versions?
- How can quadrants be linked?
- Quadrants in FCS Express versions 7.20 / 7.24 and later
- Why does the Quadrants Options window appear when I open an older layout in version 7.24?
- Why have percentages reported by quadrants changed after updating to FCS Express version 7.20.20?
-
-
Data Analysis
-
- Caveats when using Biexponential Scaling with automatic Below Zero parameter detection in the presence of outliers.
- How can I create a merged data with equally-sized downsampled samples?
- How can I do pre-processing for high-dimensional data analysis?
- How can I explore tSNE/UMAP plots?
- How do I use SPADE?
- What is FlowSOM?
- What is T-SNE?
- What is UMAP?
-
- Can FCS Express integrate Python scripts?
- Can I use the FlowAI script in FCS Express?
- Can I use the FlowClean R Script with FCS Express?
- How can I create a matrix of autofluorescence, and import autofluorescence into EasyPanel?
- How can I recreate ratiometric data acquired in FACSDiva?
- How do I use R Integration with FCS Express?
- How does FCS Express implement software compensation?
- If my data does not have a Time parameter, can I create one?
- What is compensation?
- What is the compensation workflow in FCS Express?
- When acquiring spectral data, should my single-stained controls be "as bright or brighter" than my fully-stained sample?
-
- Can a set of quadrants be both percentile and floating?
- Can I customize the display of my data from different instruments?
- Can I disable the live updating feature?
- How can I display all of my detectors for my Cytek data?
- How can I set FCS Express so my FCS 3.0 biexponential data looks the same as it did in the BD FACSDiva software?
- How do I display Summit data in FCS Express as it appears in the Summit Software?
- How do I fix the biexponential axes on a plot?
- How do I rescale CytoFLEX and DxFLEX data so it displays as it did at acquisition?
- How do I update my density and contour plots created in Version 4 to use the newest color palette?
- What are resolution options?
- What is Biexponential and Hyperlog Scaling?
- What is the best way to set FCS Express to display FCS 3.0 data from FACSDiva on a 4 decade log scale?
- Where can I get more information regarding DNA analysis using the Multicycle AV?
- Why can’t I change my plot axis labels from the Name keyword to the Stain keyword?
- Why do I see a warning message when inserting a Spectrum Plot?
- Why do my dot plots appear sparse and blocky?
- Why is the text on the right most label cut off my plot?
- Show all articles ( 2 ) Collapse Articles
-
- Can I create an output file that contains the same plot from each data file on a single page?
- Can I export spectral data to the FCS format?
- How can I successfully export a GatingML file?
- How do the batch processing run modes differ, and why would I use them?
- Why do I get an “Old format or invalid type library” error when using Microsoft excel during batch analysis?
-
- How are statistics in FCS Express calculated compared to how they are calculated in BD FACSDiva?
- How can I display my statistical data in Scientific Notation?
- How do I calculate EC/IC Anything?
- What is “Stain Index” and how do I calculate it with FCS Express?
- What is MFI (Mean or Median Fluorescence Intensity) and how do I calculate it in FCS Express?
- Why have percentages reported by quadrants changed after updating to FCS Express version 7.20.20?
- Why is the Geometric Mean being reported as NaN or ##ERROR##?
-
-
Image Cytometry
-
- How do I adjust the axes to display small particle data from Amnis CellStream?
- How do I choose which images and parameters to view in a Data Grid?
- How do I export/save data from IDEAS software and load it in FCS Express?
- How do I make my images in the data grid larger?
- How do I pseudo-color images in a data grid?
- How do I work with Amnis derived image cytometry data in FCS Express?
-
- Can I display heat maps with my Image Cytometry data?
- Can I work with data from PerkinElmer Instruments?
- Do you offer 21 CFR Part 11 compliance options for the Image Cytometry Version?
- Do you offer image segmentation or image analysis?
- How can Attune™ CytPix data sets with images (.ACS files) be merged for high dimensional analysis?
- How do I use CellProfiler Data with FCS Express?
- How do I use ImageJ with FCS Express?
- What file formats are compatible with FCS Express Image Cytometry?
- Where can I find Nexcelom Resources and Applications?
-
-
FCS Express on Mac
-
Upgrading FCS Express
-
- Can different versions of FCS Express exist on the same computer?
- How can I view and convert my V3 layouts to FCS Express 7?
- How do I import my version 3 security databases into newer versions of FCS Express?
- How do I update Density Plots created in Version 4?
- Is there an upgrade discount from earlier versions of FCS Express?
- Version 4 Internet Dongle Retirement
- Why are my density plots from V3 not displayed correctly in later versions?
- Why are there fewer outlier dots on my FCS Express 5 and later density plots than in V4?
-
Clinical & Validation Ready
-
- Can I get more information regarding the Add-Ons that can be purchased with a license?
- Can I lock my template based on an electronic signature?
- Does FCS Express have any features to help meet 21 CFR Part 11 compliance?
- What is the difference between the different types of Users that are available with a Security and Logging license?
- What is the difference between the Logging option and System Level Audit Trails?
 Explore the Scientific R&D Software
Explore the Scientific R&D Software