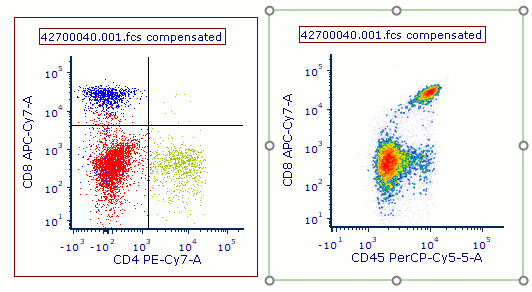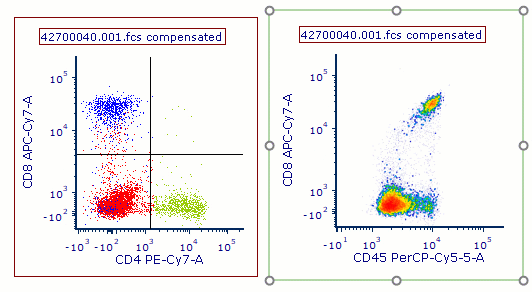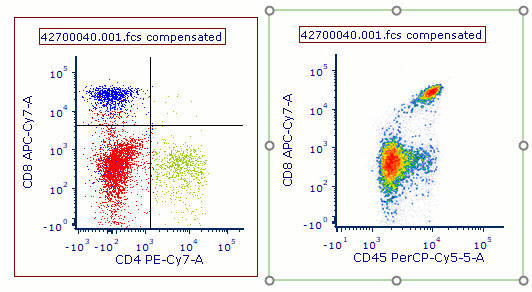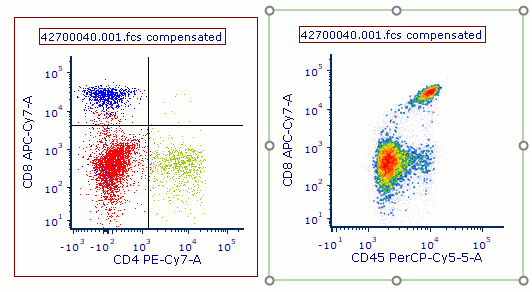
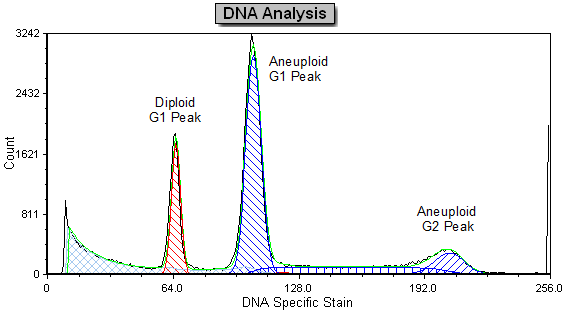
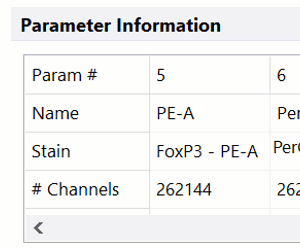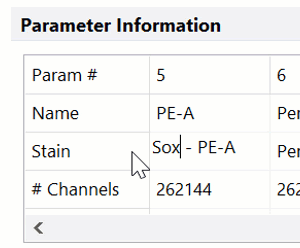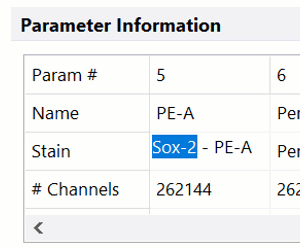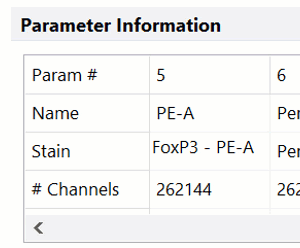
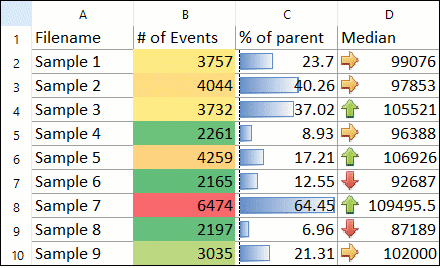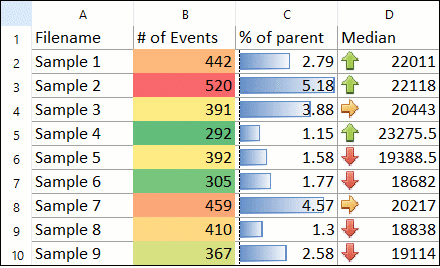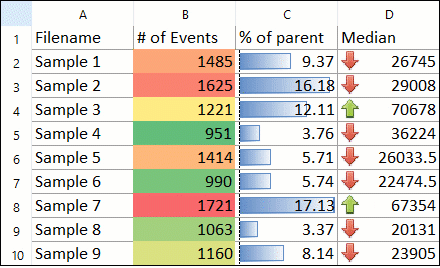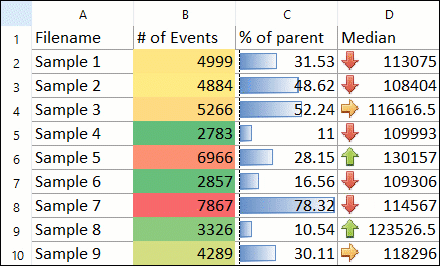
DNA Analysis with Multicycle AV
Multicycle AV™ is included free with each FCS Express license. Multicycle will allow users to fit up to 3 cell cycles with background, debris, and aggregate correction within a sample.
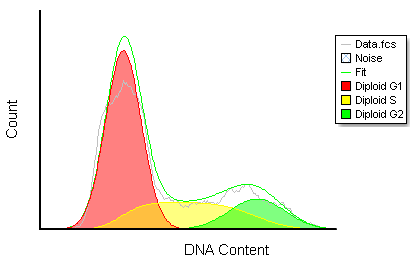
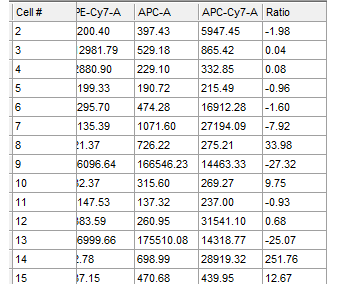
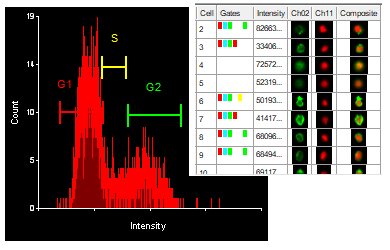
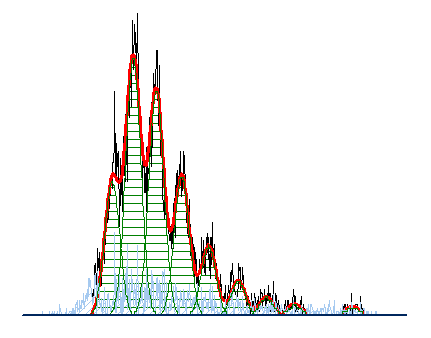
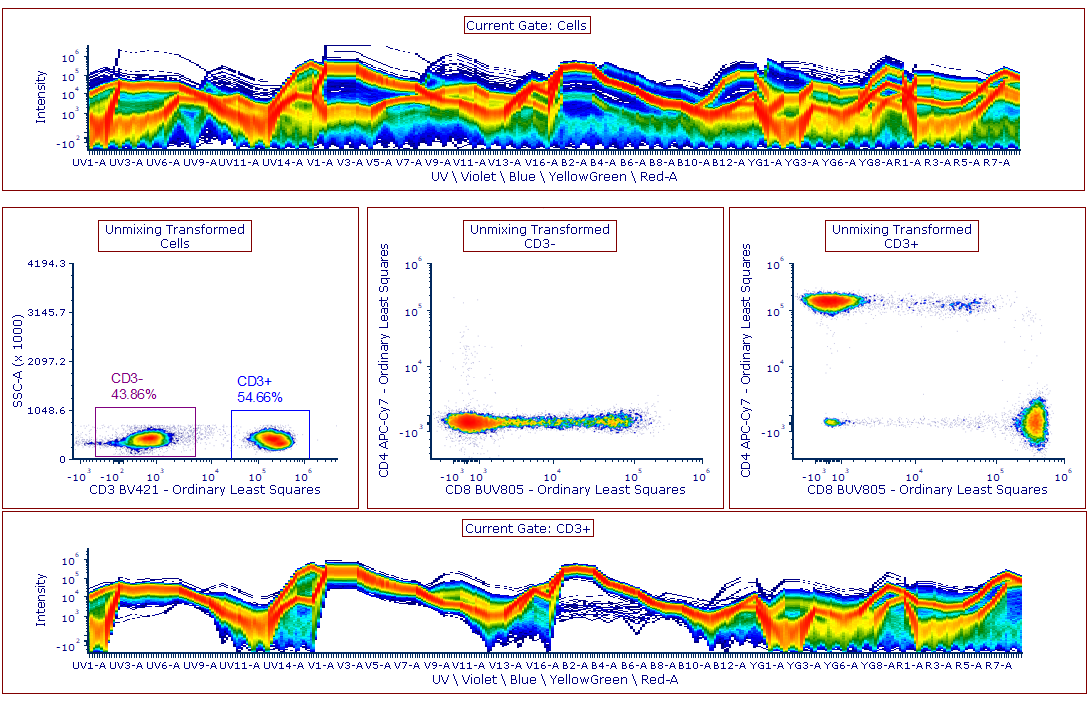
Often when trying to analyze DNA histograms for cell cycle or DNA analysis, the use of simple region markers on the histogram is not adequate. Especially if the DNA histogram exhibits a lot of debris from poor sample preparation and/or there are multiple DNA populations with different DNA content in the same histogram. When these conditions arise, it is necessary to use a powerful mathematical model to remove the debris component and deconvolute the multiple peaks in the histogram.
The MultiCycle AV DNA tool in FCS Express addresses this problem in a seamless manner. The Dean/Jett method, which is considered the gold standard for cell cycle modeling, was first proposed in 1974. In 1980, a modification by Fox was added to the Dean/Jett method to better fit data with complication S phase distributions. Multicycle AV uses the Dean/Jett/Fox model.
In addition, sophisticated debris subtraction models developed over the past decade by leaders in the field of cell cycle analysis, Rabinovitch & Bagwell, are applied to the debris component. This results in accurate, simple-to-understand cell cycle results from even the most complicated DNA histogram.
FCS Express is pleased to announce that De Novo Software has partnered with Phoenix Flow Systems and the University of Washington to combine the comprehensive DNA analysis capabilities of Multicycle AV with FCS Express.
You can now obtain the best of both worlds: the ease of use, flexible data analysis, and flexible reporting of FCS Express as well the complete suite of DNA modeling tools available in Multicycle for cell cycle analysis.

The DNA fitting engine in Multicycle has been used for over a decade by researchers all over the world and is described in several publications:
- Dean P. and Jett J., Mathematical analysis of DNA distribution derived from flow micro-fluorimetry., Cell Biology 60: 523, 1974.
- Fox M. A Model for the Computer Analysis of Synchronous DNA Distributions Obtained by Flow Cytometry. Cytometry Volume 1, No.1 71:77, 1980.
- Rabinovitch, P.S. DNA content histogram and cell-cycle analysis. Methods in Cell Biology. 41:263-296, 1994.
- Rabinovitch, P.S. Practical considerations for DNA content and cell cycle analysis. In: Clinical Flow Cytometry: Principles and Applications (Bauer, K.D., Duque, R.E., and Shankey, T.V., eds.) Williams and Wilkins, Baltimore, pp. 117‑142, 1992.
- Rabinovitch, P.S. DNA Histogram Analysis. In: Cell Growth and Division, 2nd Edition. (G. Studinski, ed.). Oxford University Press, Oxford, 1996.
- Kallioniemi, O-P., Visakorpi, T., Holli, K., Isola, J.J., and Rabinovitch, P.S. Automated peak detection and cell cycle analysis of flow cytometric histograms. Cytometry 16:250-255, 1994